Structure and mechanism of a Na+-independent amino acid transporter
- PMID: 19608859
- PMCID: PMC2851542
- DOI: 10.1126/science.1176088
Structure and mechanism of a Na+-independent amino acid transporter
Abstract
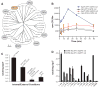
Amino acid, polyamine, and organocation (APC) transporters are secondary transporters that play essential roles in nutrient uptake, neurotransmitter recycling, ionic homeostasis, and regulation of cell volume. Here, we present the crystal structure of apo-ApcT, a proton-coupled broad-specificity amino acid transporter, at 2.35 angstrom resolution. The structure contains 12 transmembrane helices, with the first 10 consisting of an inverted structural repeat of 5 transmembrane helices like the leucine transporter LeuT. The ApcT structure reveals an inward-facing, apo state and an amine moiety of lysine-158 located in a position equivalent to the sodium ion site Na2 of LeuT. We propose that lysine-158 is central to proton-coupled transport and that the amine group serves the same functional role as the Na2 ion in LeuT, thus demonstrating common principles among proton- and sodium-coupled transporters.
Figures

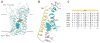
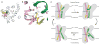
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

