Functional role for the conformationally mobile phenylalanine 223 in the reaction of methylenetetrahydrofolate reductase from Escherichia coli
- PMID: 19610625
- PMCID: PMC2805098
- DOI: 10.1021/bi9007325
Functional role for the conformationally mobile phenylalanine 223 in the reaction of methylenetetrahydrofolate reductase from Escherichia coli
Abstract
The flavoprotein methylenetetrahydrofolate reductase from Escherichia coli catalyzes the reduction of 5,10-methylenetetrahydrofolate (CH(2)-H(4)folate) by NADH via a ping-pong reaction mechanism. Structures of the reduced enzyme in complex with NADH and of the oxidized Glu28Gln enzyme in complex with CH(3)-H(4)folate [Pejchal, R., Sargeant, R., and Ludwig, M. L. (2005) Biochemistry 44, 11447-11457] have revealed Phe223 as a conformationally mobile active site residue. In the NADH complex, the NADH adopts an unusual hairpin conformation and is wedged between the isoalloxazine ring of the FAD and the side chain of Phe223. In the folate complex, Phe223 swings out from its position in the NADH complex to stack against the p-aminobenzoate ring of the folate. Although Phe223 contacts each substrate in E. coli MTHFR, this residue is not invariant; for example, a leucine occurs at this site in the human enzyme. To examine the role of Phe223 in substrate binding and catalysis, we have constructed mutants Phe223Ala and Phe223Leu. As predicted, our results indicate that Phe223 participates in the binding of both substrates. The Phe223Ala mutation impairs NADH and CH(2)-H(4)folate binding each 40-fold yet slows catalysis of both half-reactions less than 2-fold. Affinity for CH(2)-H(4)folate is unaffected by the Phe223Leu mutation, and the variant catalyzes the oxidative half-reaction 3-fold faster than the wild-type enzyme. Structures of ligand-free Phe223Leu and Phe223Leu/Glu28Gln MTHFR in complex with CH(3)-H(4)folate have been determined at 1.65 and 1.70 A resolution, respectively. The structures show that the folate is bound in a catalytically competent conformation, and Leu223 undergoes a conformational change similar to that observed for Phe223 in the Glu28Gln-CH(3)-H(4)folate structure. Taken together, our results suggest that Leu may be a suitable replacement for Phe223 in the oxidative half-reaction of E. coli MTHFR.
Figures

 ), Phe223Ala, (
), Phe223Ala, (
 ), and wild-type, (
), and wild-type, ( ) enzymes (30 μM) in 50 mM potassium phosphate (pH 7.2) buffer containing 0.3 mM EDTA and 10% glycerol. For all enzymes, wavelength of maximal absorbance is 447 nm and molar extinction coefficient is 14,300 M−1cm−1.
) enzymes (30 μM) in 50 mM potassium phosphate (pH 7.2) buffer containing 0.3 mM EDTA and 10% glycerol. For all enzymes, wavelength of maximal absorbance is 447 nm and molar extinction coefficient is 14,300 M−1cm−1.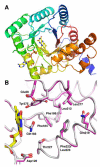

 ), 100 (
), 100 ( ), 200 (
), 200 (
 ), 300 (
), 300 ( ), 500 (
), 500 ( ), 800 (
), 800 ( ), 1500 (
), 1500 ( ), and 2000 (
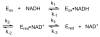
), and 2000 ( ) μM NADH (concentrations after mixing). Stopped-flow reaction traces were monitored at 450 nm. The reaction was monophasic. (Inset) Dependence of the observed rate constant for reduction on NADH concentration. The data were fit to a hyperbolic equation (eq 4), which yielded an apparent Kd of 440 ± 20 μM for NADH and a maximum rate constant for reduction (k2’, according to Scheme 2) of 35 ± 1 s−1. Reaction conditions were 50 mM potassium phosphate (pH 7.2) buffer containing 0.3 mM EDTA and 10% glycerol at 25 °C.
) μM NADH (concentrations after mixing). Stopped-flow reaction traces were monitored at 450 nm. The reaction was monophasic. (Inset) Dependence of the observed rate constant for reduction on NADH concentration. The data were fit to a hyperbolic equation (eq 4), which yielded an apparent Kd of 440 ± 20 μM for NADH and a maximum rate constant for reduction (k2’, according to Scheme 2) of 35 ± 1 s−1. Reaction conditions were 50 mM potassium phosphate (pH 7.2) buffer containing 0.3 mM EDTA and 10% glycerol at 25 °C.
 ), 25 (
), 25 (
 ), 50 (
), 50 ( ), 100 (
), 100 ( ), 200 (
), 200 ( ), and 300 μM (
), and 300 μM ( ) CH2-H4folate (concentrations after mixing). Stopped-flow reaction traces, monitored at 450 nm, were monophasic. (Inset) Dependence of the observed rate constant on CH2-H4folate concentration. A fit of the data to eq 4 yielded an apparent Kd for CH2-H4folate of 9 ± 2 μM and a maximal rate constant for oxidation (k5’, according to Scheme 3) of 34 ± 1 s−1. Reaction conditions were 50 mM potassium phosphate (pH 7.2) buffer containing 0.3 mM EDTA and 10% glycerol at 25 °C.
) CH2-H4folate (concentrations after mixing). Stopped-flow reaction traces, monitored at 450 nm, were monophasic. (Inset) Dependence of the observed rate constant on CH2-H4folate concentration. A fit of the data to eq 4 yielded an apparent Kd for CH2-H4folate of 9 ± 2 μM and a maximal rate constant for oxidation (k5’, according to Scheme 3) of 34 ± 1 s−1. Reaction conditions were 50 mM potassium phosphate (pH 7.2) buffer containing 0.3 mM EDTA and 10% glycerol at 25 °C.
 ), 100 (
), 100 (
 ), 190 (
), 190 ( ), 385 (
), 385 ( ), 580 (
), 580 ( ), and 980 (
), and 980 ( ) μM CH2-H4folate (concentrations after mixing). Stopped-flow reaction traces, monitored at 450 nm, were biphasic. The fast and slow phases accounted for 84%, and 16%, respectively, of the total observed absorbance change. (Inset). Dependence of the slow phase on CH2-H4folate concentration. A hyperbolic fit of the data gave an apparent Kd of 500 ± 230 μM for CH2-H4folate and a maximum rate constant of 4.5 ± 1.1 s−1. Reaction conditions were 50 mM potassium phosphate (pH 7.2) buffer containing 0.3 mM EDTA and 10% glycerol at 25 °C.
) μM CH2-H4folate (concentrations after mixing). Stopped-flow reaction traces, monitored at 450 nm, were biphasic. The fast and slow phases accounted for 84%, and 16%, respectively, of the total observed absorbance change. (Inset). Dependence of the slow phase on CH2-H4folate concentration. A hyperbolic fit of the data gave an apparent Kd of 500 ± 230 μM for CH2-H4folate and a maximum rate constant of 4.5 ± 1.1 s−1. Reaction conditions were 50 mM potassium phosphate (pH 7.2) buffer containing 0.3 mM EDTA and 10% glycerol at 25 °C.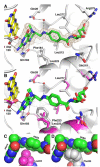


References
-
- Jacques PF, Bostom AG, Williams RR, Ellison RC, Eckfeldt JH, Rosenberg IH, Selhub J, Rozen R. Relation between folate status, a common mutation in methylenetetrahydrofolate reductase, and plasma homocysteine concentrations. Circulation. 1996;93:7–9. - PubMed
-
- Chen Z, Karaplis AC, Ackerman SL, Pogribny IP, Melnyk S, Lussier-Cacan S, Chen MF, Pai A, John SWM, Smith RS, Bottiglieri T, Bagley P, Selhub J, Rudnicki MA, James SJ, Rozen R. Mice deficient in methylenetetrahydrofolate reductase exhibit hyperhomocysteinemia and decreased methylation capacity, with neuropathology and aortic lipid deposition. Hum. Mol. Genet. 2001;10:433–443. - PubMed
-
- Refsum H, Ueland PM, Nygard O, Vollset SE. Homocysteine and Cardiovascular Disease. Annu. Rev. Medicine. 1998;49:31–62. - PubMed
-
- McCaddon A, Davies G, Hudson P, Tandy S, Cattell H. Total serum homocysteine in senile dementia of alzheimer type. Int. J. Geriatr. Psychiatry. 1998;13:235–239. - PubMed
-
- Clarke R, Smith AD, Jobst KA, Refsum H, Sutton L, Ueland PM. Folate, vitamin B12, and serum total homocysteine levels in confirmed Alzheimer disease. Arch. Neurol. 1998;55:1449–1455. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

