Evolution of a subtilisin-like protease gene family in the grass endophytic fungus Epichloë festucae
- PMID: 19615101
- PMCID: PMC2717940
- DOI: 10.1186/1471-2148-9-168
Evolution of a subtilisin-like protease gene family in the grass endophytic fungus Epichloë festucae
Abstract
Background: Subtilisin-like proteases (SLPs) form a superfamily of enzymes that act to degrade protein substrates. In fungi, SLPs can play either a general nutritive role, or may play specific roles in cell metabolism, or as pathogenicity or virulence factors.
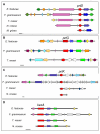
Results: Fifteen different genes encoding SLPs were identified in the genome of the grass endophytic fungus Epichloë festucae. Phylogenetic analysis indicated that these SLPs belong to four different subtilisin families: proteinase K, kexin, pyrolysin and subtilisin. The pattern of intron loss and gain is consistent with this phylogeny. E. festucae is exceptional in that it contains two kexin-like genes. Phylogenetic analysis in Hypocreales fungi revealed an extensive history of gene loss and duplication.
Conclusion: This study provides new insights into the evolution of the SLP superfamily in filamentous fungi.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

