Comparative analysis of conditional reporter alleles in the developing embryo and embryonic nervous system
- PMID: 19616131
- PMCID: PMC2855890
- DOI: 10.1016/j.gep.2009.07.007
Comparative analysis of conditional reporter alleles in the developing embryo and embryonic nervous system
Abstract
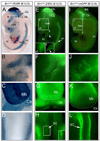
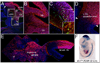
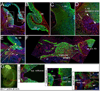
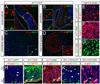
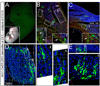
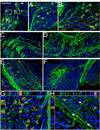
A long-standing problem in development is understanding how progenitor cells transiently expressing genes contribute to complex anatomical and functional structures. In the developing nervous system an additional level of complexity arises when considering how cells of distinct lineages relate to newly established neural circuits. To address these problems, we used both cumulative marking with Cre/loxP and Genetic Inducible Fate Mapping (GIFM), which permanently and heritably marks small populations of progenitors and their descendants with fine temporal control using CreER/loxP. A key component used in both approaches is a conditional phenotyping allele that has the potential to be expressed in all cell types, but is quiescent because of a loxP flanked Stop sequence, which precedes a reporter allele. Upon recombination, the resulting phenotyping allele is 'turned on' and then constitutively expressed. Thus, the reporter functions as a high fidelity genetic lineage tracer in vivo. Currently there is an array of reporter alleles that can be used in marking strategies, but their recombination efficiency and applicability to a wide array of tissues has not been thoroughly described. To assess the recombination/marking potential of the reporters, we utilized CreER(T) under the control of a Wnt1 transgene (Wnt1-CreER(T)) as well as a cumulative, non-inducible En1(Cre) knock-in line in combination with three different reporters: R26R (LacZ reporter), Z/EG (EGFP reporter), and Tau-Lox-STOP-Lox-mGFP-IRES-NLS-LacZ (membrane-targeted GFP/nuclear LacZ reporter). We marked the Wnt1 lineage using each of the three reporters at embryonic day (E) 8.5 followed by analysis at E10.0, E12.5, and in the adult. We also compared cumulative marking of cells with a history of En1 expression at the same stages. We evaluated the reporters by whole-mount and section analysis and ascertained the strengths and weaknesses of each of the reporters. Comparative analysis with the reporters elucidated complexities of how the Wnt1 and En1 lineages contribute to developing embryos and to axonal projection patterns of neurons derived from these lineages.
Figures




References
-
- Ahn S, Joyner AL. Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell. 2004;118:505–516. - PubMed
-
- Ahn S, Joyner AL. In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature. 2005;437:894–897. - PubMed
-
- Davidson D, Graham E, Sime C, Hill R. A gene with sequence similarity to Drosophila engrailed is expressed during the development of the neural tube and vertebrae in the mouse. Development. 1988;104:305–316. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

