Variations on a theme: eukaryotic Y-family DNA polymerases
- PMID: 19616647
- PMCID: PMC2846237
- DOI: 10.1016/j.bbapap.2009.07.004
Variations on a theme: eukaryotic Y-family DNA polymerases
Abstract
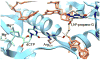
Most classical DNA polymerases, which function in normal DNA replication and repair, are unable to synthesize DNA opposite damage in the template strand. Thus in order to replicate through sites of DNA damage, cells are equipped with a variety of nonclassical DNA polymerases. These nonclassical polymerases differ from their classical counterparts in at least two important respects. First, nonclassical polymerases are able to efficiently incorporate nucleotides opposite DNA lesions while classical polymerases are generally not. Second, nonclassical polymerases synthesize DNA with a substantially lower fidelity than do classical polymerases. Many nonclassical polymerases are members of the Y-family of DNA polymerases, and this article focuses on the mechanisms of the four eukaryotic members of this family: polymerase eta, polymerase kappa, polymerase iota, and the Rev1 protein. We discuss the mechanisms of these enzymes at the kinetic and structural levels with a particular emphasis on how they accommodate damaged DNA substrates. Work over the last decade has shown that the mechanisms of these nonclassical polymerases are fascinating variations of the mechanism of the classical polymerases. The mechanisms of polymerases eta and kappa represent rather minor variations, while the mechanisms of polymerase iota and the Rev1 protein represent rather major variations. These minor and major variations all accomplish the same goal: they allow the nonclassical polymerases to circumvent the problems posed by the template DNA lesion.
Copyright (c) 2010 Elsevier B.V. All rights reserved.
Figures



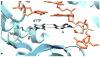
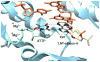
References
-
- Kunkel TA, Bebenek R. DNA replication fidelity. Annual Review of Biochemistry. 2000;69:497–529. - PubMed
-
- Doublie S, Ellenberger T. The mechanism of action of T7 DNA polymerase. Current Opinion in Structural Biology. 1998;8:704–712. - PubMed
-
- Doublie S, Sawaya MR, Ellenberger T. An open and closed case for all polymerases. Structure with Folding & Design. 1999;7:R31–R35. - PubMed
-
- Brautigam CA, Steitz TA. Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Current Opinion in Structural Biology. 1998;8:54–63. - PubMed
-
- Steitz TA. DNA polymerases: Structural diversity and common mechanisms. Journal of Biological Chemistry. 1999;274:17395–17398. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

