SNPs in human miRNA genes affect biogenesis and function
- PMID: 19617315
- PMCID: PMC2743066
- DOI: 10.1261/rna.1560209
SNPs in human miRNA genes affect biogenesis and function
Abstract
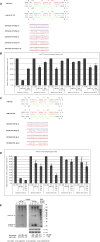
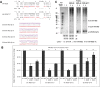
MicroRNAs (miRNAs) are 21-25-nucleotide-long, noncoding RNAs that are involved in translational regulation. Most miRNAs derive from a two-step sequential processing: the generation of pre-miRNA from pri-miRNA by the Drosha/DGCR8 complex in the nucleus, and the generation of mature miRNAs from pre-miRNAs by the Dicer/TRBP complex in the cytoplasm. Sequence variation around the processing sites, and sequence variations in the mature miRNA, especially the seed sequence, may have profound affects on miRNA biogenesis and function. In the context of analyzing the roles of miRNAs in Schizophrenia and Autism, we defined at least 24 human X-linked miRNA variants. Functional assays were developed and performed on these variants. In this study we investigate the affects of single nucleotide polymorphisms (SNPs) on the generation of mature miRNAs and their function, and report that naturally occurring SNPs can impair or enhance miRNA processing as well as alter the sites of processing. Since miRNAs are small functional units, single base changes in both the precursor elements as well as the mature miRNA sequence may drive the evolution of new microRNAs by altering their biological function. Finally, the miRNAs examined in this study are X-linked, suggesting that the mutant alleles could be determinants in the etiology of diseases.
Figures



References
-
- Abelson JF, Kwan KY, O'Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin MR, Gunel M, et al. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science. 2005;310:317–320. - PubMed
-
- Arisawa T, Tahara T, Shibata T, Nagasaka M, Nakamura M, Kamiya Y, Fujita H, Hasegawa S, Takagi T, Wang FY, et al. A polymorphism of microRNA 27a genome region is associated with the development of gastric mucosal atrophy in Japanese male subjects. Dig Dis Sci. 2007;52:1691–1697. - PubMed
-
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
