Processive endoglucanases mediate degradation of cellulose by Saccharophagus degradans
- PMID: 19617364
- PMCID: PMC2737977
- DOI: 10.1128/JB.00481-09
Processive endoglucanases mediate degradation of cellulose by Saccharophagus degradans
Abstract
Bacteria and fungi are thought to degrade cellulose through the activity of either a complexed or a noncomplexed cellulolytic system composed of endoglucanases and cellobiohydrolases. The marine bacterium Saccharophagus degradans 2-40 produces a multicomponent cellulolytic system that is unusual in its abundance of GH5-containing endoglucanases. Secreted enzymes of this bacterium release high levels of cellobiose from cellulosic materials. Through cloning and purification, the predicted biochemical activities of the one annotated cellobiohydrolase Cel6A and the GH5-containing endoglucanases were evaluated. Cel6A was shown to be a classic endoglucanase, but Cel5H showed significantly higher activity on several types of cellulose, was the highest expressed, and processively released cellobiose from cellulosic substrates. Cel5G, Cel5H, and Cel5J were found to be members of a separate phylogenetic clade and were all shown to be processive. The processive endoglucanases are functionally equivalent to the endoglucanases and cellobiohydrolases required for other cellulolytic systems, thus providing a cellobiohydrolase-independent mechanism for this bacterium to convert cellulose to glucose.
Figures
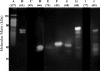
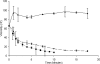

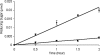

References
-
- Bayer, E. A., H. Chanzy, R. Lamed, and Y. Shoham. 1998. Cellulose, cellulases and cellulosomes. Curr. Opin. Struct. Biol. 8548-557. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

