Identification of a selective nuclear import signal in adenosine deaminases acting on RNA
- PMID: 19617375
- PMCID: PMC2761270
- DOI: 10.1093/nar/gkp599
Identification of a selective nuclear import signal in adenosine deaminases acting on RNA
Abstract
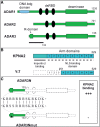
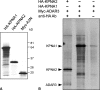
The adenosine deaminases acting on RNA (ADARs) comprise a family of RNA editing enzymes that selectively modify single codons within RNA primary transcripts with often profound impact on protein function. Little is known about the mechanisms that regulate nuclear RNA editing activity. Editing levels show cell-type specific and developmental modulation that does not strictly coincide with observed expression levels of ADARs. Here, we provide evidence for a molecular mechanism that might control nuclear import of specific ADARs and, in turn, nuclear RNA editing. We identify an in vivo ADAR3 interaction partner, importin alpha 1 (KPNA2) that specifically recognizes an arginine-rich ADAR3 sequence motif and show that it acts as a functional nuclear localization sequence. Furthermore, whereas KPNA2, but not KPNA1 or KNPA3, recognizes the ADAR3 NLS, we observe the converse binding specificity with ADAR2. Interestingly, alternative splicing of ADAR2 pre-mRNA introduces an ADAR3-like NLS that alters the interaction profile with the importins. Thus, in vivo RNA editing might be regulated, in part, through controlled subcellular localization of ADARs, which in turn is governed by the coordinated local expression of importin alpha proteins and ADAR protein variants.
Figures


References
-
- Gommans WM, Dupuis DE, McCane JE, Tatalias NE, Maas S. In: DNA RNA Editing. Smith H, editor. New York: Wiley & Sons, Inc; 2008. pp. 3–30.
-
- Maydanovych O, Beal PA. Breaking the central dogma by RNA editing. Chem. Rev. 2006;106:3397–3411. - PubMed
-
- Rueter SM, Dawson TR, Emeson RB. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

