Energetics of glutamate receptor ligand binding domain dimer assembly are modulated by allosteric ions
- PMID: 19617541
- PMCID: PMC2718364
- DOI: 10.1073/pnas.0904175106
Energetics of glutamate receptor ligand binding domain dimer assembly are modulated by allosteric ions
Abstract
The activity of many ligand-gated ion channels and cell surface receptors is modulated by small molecules and ions, but an understanding of the underlying molecular mechanisms is scarce. For kainate, but not AMPA subtype glutamate receptors, the binding of Na(+) and Cl(-) ions to discrete, electrostatically coupled sites in the extracellular ligand binding domain (LBD) dimer assembly regulates the rate of entry into the desensitized state, which occurs when the dimer interface ruptures and the channel closes. Studies on glutamate receptors have defined the LBD dimer assembly as a key functional unit that controls activation and desensitization. Here we use analytical ultracentrifugation to probe the energetic effects of allosteric ions on kainate receptor dimer stability in solution, using a GluR6 mutant that desensitizes slowly. Our results show that sodium and chloride ions modulate kainate receptor dimer affinity as much as 50-fold, and that removal of either Cl(-) or Na(+) disrupts the dimer. The applicability of a similar allosteric mechanism for modulation of delta2 glutamate receptors by Ca(2+) was also tested. Our results indicate that ions can contribute substantial free energy to active state stabilization in both these receptors, and provide quantitative measurements of the energetic consequences of allosteric ion binding to a ligand-gated ion channel.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
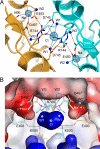

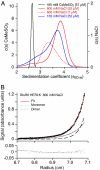
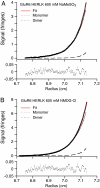
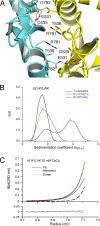
References
-
- Changeux JP, Edelstein SJ. Allosteric receptors after 30 years. Neuron. 1998;21:959–980. - PubMed
-
- Sun Y, et al. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. - PubMed
-
- Silberberg SD, Li M, Swartz KJ. Ivermectin interaction with transmembrane helices reveals widespread rearrangements during opening of P2X receptor channels. Neuron. 2007;54:263–274. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

