High frequency of hotspot mutations in core genes of Escherichia coli due to short-term positive selection
- PMID: 19617543
- PMCID: PMC2718352
- DOI: 10.1073/pnas.0906217106
High frequency of hotspot mutations in core genes of Escherichia coli due to short-term positive selection
Abstract
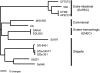
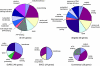
Core genes comprising the ubiquitous backbone of bacterial genomes are not subject to frequent horizontal transfer and generally are not thought to contribute to the adaptive evolution of bacterial pathogens. We determined, however, that at least one-third and possibly more than one-half of the core genes in Escherichia coli genomes are targeted by repeated replacement substitutions in the same amino acid positions-hotspot mutations. Occurrence of hotspot mutations is driven by positive selection, as their rate is significantly higher than expected by random chance alone, and neither intragenic recombination nor increased mutability can explain the observed patterns. Also, commensal E. coli strains have a significantly lower frequency of mutated genes and mutations per genome than pathogenic strains. E. coli strains causing extra-intestinal infections accumulate hotspot mutations at the highest rate, whereas the highest total number of mutated genes has been found among Shigella isolates, suggesting the pathoadaptive nature of such mutations. The vast majority of hotspot mutations are of recent evolutionary origin, implying short-term positive selection, where adaptive mutations emerge repeatedly but are not sustained in natural circulation for long. Such pattern of dynamics is consistent with source-sink model of virulence evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Ghedin E, et al. Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature. 2005;437:1162–1166. - PubMed
-
- Shastry BS. SNPs in disease gene mapping, medicinal drug development, and evolution. J Hum Genet. 2007;52:871–880. - PubMed
-
- Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol. 2004;2:414–424. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

