Long term non-invasive imaging of embryonic stem cells using reporter genes
- PMID: 19617890
- PMCID: PMC3683546
- DOI: 10.1038/nprot.2009.100
Long term non-invasive imaging of embryonic stem cells using reporter genes
Abstract
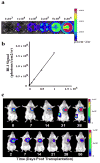
Development of non-invasive and accurate methods to track cell fate after delivery will greatly expedite transition of embryonic stem (ES) cell therapy to the clinic. In this protocol, we describe the in vivo monitoring of stem cell survival, proliferation and migration using reporter genes. We established stable ES cell lines constitutively expressing double fusion (DF; enhanced green fluorescent protein and firefly luciferase) or triple fusion (TF; monomeric red fluorescent protein, firefly luciferase and herpes simplex virus thymidine kinase (HSVtk)) reporter genes using lentiviral transduction. We used fluorescence-activated cell sorting to purify these populations in vitro, bioluminescence imaging and positron emission tomography (PET) imaging to track them in vivo and fluorescence immunostaining to confirm the results ex vivo. Unlike other methods of cell tracking, such as iron particle and radionuclide labeling, reporter genes are inherited genetically and can be used to monitor cell proliferation and survival for the lifetime of transplanted cells and their progeny.
Conflict of interest statement
The authors declare that they have no competing financial interests.
Figures



Similar articles
-
Imaging of embryonic stem cell migration in vivo.Methods Mol Biol. 2011;750:101-14. doi: 10.1007/978-1-61779-145-1_7. Methods Mol Biol. 2011. PMID: 21618086 Free PMC article.
-
Molecular imaging of embryonic stem cell misbehavior and suicide gene ablation.Cloning Stem Cells. 2007 Spring;9(1):107-17. doi: 10.1089/clo.2006.0E16. Cloning Stem Cells. 2007. PMID: 17386018
-
Transcriptional profiling of reporter genes used for molecular imaging of embryonic stem cell transplantation.Physiol Genomics. 2006 Mar 13;25(1):29-38. doi: 10.1152/physiolgenomics.00254.2005. Epub 2006 Jan 3. Physiol Genomics. 2006. PMID: 16390873
-
Clinical hurdles for the transplantation of cardiomyocytes derived from human embryonic stem cells: role of molecular imaging.Curr Opin Biotechnol. 2007 Feb;18(1):38-45. doi: 10.1016/j.copbio.2006.12.003. Epub 2006 Dec 29. Curr Opin Biotechnol. 2007. PMID: 17196814 Review.
-
Non-invasive imaging of human embryonic stem cells.Curr Pharm Biotechnol. 2010 Sep 1;11(6):685-92. doi: 10.2174/138920110792246500. Curr Pharm Biotechnol. 2010. PMID: 20497109 Free PMC article. Review.
Cited by
-
CCND2 Overexpression Enhances the Regenerative Potency of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes: Remuscularization of Injured Ventricle.Circ Res. 2018 Jan 5;122(1):88-96. doi: 10.1161/CIRCRESAHA.117.311504. Epub 2017 Oct 10. Circ Res. 2018. PMID: 29018036 Free PMC article.
-
Fast and efficient multitransgenic modification of human pluripotent stem cells.Hum Gene Ther Methods. 2014 Apr;25(2):136-53. doi: 10.1089/hgtb.2012.248. Epub 2014 Mar 21. Hum Gene Ther Methods. 2014. PMID: 24483184 Free PMC article.
-
Imaging neural stem cell graft-induced structural repair in stroke.Cell Transplant. 2013;22(5):881-92. doi: 10.3727/096368912X656144. Cell Transplant. 2013. PMID: 23044338 Free PMC article.
-
Y-27632 preconditioning enhances transplantation of human-induced pluripotent stem cell-derived cardiomyocytes in myocardial infarction mice.Cardiovasc Res. 2019 Feb 1;115(2):343-356. doi: 10.1093/cvr/cvy207. Cardiovasc Res. 2019. PMID: 30107391 Free PMC article.
-
Enhanced noninvasive imaging of oncology models using the NIS reporter gene and bioluminescence imaging.Cancer Gene Ther. 2020 Apr;27(3-4):179-188. doi: 10.1038/s41417-019-0081-2. Epub 2019 Jan 24. Cancer Gene Ther. 2020. PMID: 30674994 Free PMC article.
References
-
- Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. - PubMed
-
- Reinlib L, Field L. Cell transplantation as future therapy for cardiovascular disease?: A workshop of the National Heart, Lung, and Blood Institute. Circulation. 2000;101:E182–187. - PubMed
-
- Spergel DJ, Kruth U, Shimshek DR, Sprengel R, Seeburg PH. Using reporter genes to label selected neuronal populations in transgenic mice for gene promoter, anatomical, and physiological studies. Prog Neurobiol. 2001;63:673–686. - PubMed
-
- Massoud TF, Gambhir SS. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev. 2003;17:545–580. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

