Probing the urea dependence of residual structure in denatured human alpha-lactalbumin
- PMID: 19618277
- PMCID: PMC2728226
- DOI: 10.1007/s10858-009-9342-y
Probing the urea dependence of residual structure in denatured human alpha-lactalbumin
Abstract
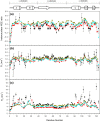
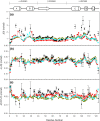
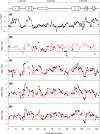
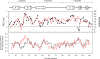
Backbone (15)N relaxation parameters and (15)N-(1)H(N) residual dipolar couplings (RDCs) have been measured for a variant of human alpha-lactalbumin (alpha-LA) in 4, 6, 8 and 10 M urea. In the alpha-LA variant, the eight cysteine residues in the protein have been replaced by alanines (all-Ala alpha-LA). This protein is a partially folded molten globule at pH 2 and has been shown previously to unfold in a stepwise non-cooperative manner on the addition of urea. (15)N R(2) values in some regions of all-Ala alpha-LA show significant exchange broadening which is reduced as the urea concentration is increased. Experimental RDC data are compared with RDCs predicted from a statistical coil model and with bulkiness, average area buried upon folding and hydrophobicity profiles in order to identify regions of non-random structure. Residues in the regions corresponding to the B, D and C-terminal 3(10) helices in native alpha-LA show R(2) values and RDC data consistent with some non-random structural propensities even at high urea concentrations. Indeed, for residues 101-106 the residual structure persists in 10 M urea and the RDC data suggest that this might include the formation of a turn-like structure. The data presented here allow a detailed characterization of the non-cooperative unfolding of all-Ala alpha-LA at higher concentrations of denaturant and complement previous studies which focused on structural features of the molten globule which is populated at lower concentrations of denaturant.
Figures

Similar articles
-
Alpha-lactalbumin forms a compact molten globule in the absence of disulfide bonds.Nat Struct Biol. 1999 Oct;6(10):948-52. doi: 10.1038/13318. Nat Struct Biol. 1999. PMID: 10504730
-
Local and global cooperativity in the human alpha-lactalbumin molten globule.J Mol Biol. 2004 Apr 16;338(1):149-58. doi: 10.1016/j.jmb.2004.02.045. J Mol Biol. 2004. PMID: 15050830
-
Pressure-induced unfolding of the molten globule of all-Ala alpha-lactalbumin.Protein Sci. 2003 Jan;12(1):66-72. doi: 10.1110/ps.0221303. Protein Sci. 2003. PMID: 12493829 Free PMC article.
-
Peptide models of local and long-range interactions in the molten globule state of human alpha-lactalbumin.J Mol Biol. 1998;283(1):279-91. doi: 10.1006/jmbi.1998.2099. J Mol Biol. 1998. PMID: 9761690
-
Using nuclear magnetic resonance spectroscopy to study molten globule states of proteins.Methods. 2004 Sep;34(1):121-32. doi: 10.1016/j.ymeth.2004.03.009. Methods. 2004. PMID: 15283921 Review.
Cited by
-
A Combined NMR and SAXS Analysis of the Partially Folded Cataract-Associated V75D γD-Crystallin.Biophys J. 2017 Mar 28;112(6):1135-1146. doi: 10.1016/j.bpj.2017.02.010. Biophys J. 2017. PMID: 28355541 Free PMC article.
-
A topical issue: NMR investigations of molecular dynamics.J Biomol NMR. 2009 Sep;45(1-2):1-4. doi: 10.1007/s10858-009-9345-8. J Biomol NMR. 2009. PMID: 19669621 Free PMC article. No abstract available.
-
α-Lactalbumin, Amazing Calcium-Binding Protein.Biomolecules. 2020 Aug 20;10(9):1210. doi: 10.3390/biom10091210. Biomolecules. 2020. PMID: 32825311 Free PMC article. Review.
References
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1002/prot.340020207', 'is_inner': False, 'url': 'https://doi.org/10.1002/prot.340020207'}, {'type': 'PubMed', 'value': '3447171', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/3447171/'}]}
- Abraham DJ, Leo AJ (1987) Extension of the fragment method to calculate amino acid zwitterion and side chain partition coefficients. Proteins 2:130–152 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1021/bi0120796', 'is_inner': False, 'url': 'https://doi.org/10.1021/bi0120796'}, {'type': 'PubMed', 'value': '11863448', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/11863448/'}]}
- Ackerman MS, Shortle D (2002a) Molecular alignment of denatured states of staphylococcal nuclease with strained polyacrylamide gels and surfactant liquid crystalline phases. Biochemistry 41:3089–3095 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1021/bi020511t', 'is_inner': False, 'url': 'https://doi.org/10.1021/bi020511t'}, {'type': 'PubMed', 'value': '12427042', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/12427042/'}]}
- Ackerman MS, Shortle D (2002b) Robustness of the long-range structure in denatured staphylococcal nuclease to changes in amino acid sequence. Biochemistry 41:13791–13797 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1110/ps.03164403', 'is_inner': False, 'url': 'https://doi.org/10.1110/ps.03164403'}, {'type': 'PMC', 'value': 'PMC2366913', 'is_inner': False, 'url': 'https://pmc.ncbi.nlm.nih.gov/articles/PMC2366913/'}, {'type': 'PubMed', 'value': '14500871', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/14500871/'}]}
- Alexandrescu AT, Kammerer RA (2003) Structure and disorder in the ribonuclease S-peptide probed by NMR residual dipolar couplings. Protein Sci 12:2132–2140 - PMC - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1016/S0065-3233(00)53005-8', 'is_inner': False, 'url': 'https://doi.org/10.1016/s0065-3233(00)53005-8'}, {'type': 'PubMed', 'value': '10751946', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/10751946/'}]}
- Arai M, Kuwajima K (2000) Role of the molten globule state in protein folding. Adv Prot Chem 53:209–282 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

