Arsenophonus, an emerging clade of intracellular symbionts with a broad host distribution
- PMID: 19619300
- PMCID: PMC2724383
- DOI: 10.1186/1471-2180-9-143
Arsenophonus, an emerging clade of intracellular symbionts with a broad host distribution
Abstract
Background: The genus Arsenophonus is a group of symbiotic, mainly insect-associated bacteria with rapidly increasing number of records. It is known from a broad spectrum of hosts and symbiotic relationships varying from parasitic son-killers to coevolving mutualists.The present study extends the currently known diversity with 34 samples retrieved mainly from hippoboscid (Diptera: Hippoboscidae) and nycteribiid (Diptera: Nycteribiidae) hosts, and investigates phylogenetic relationships within the genus.
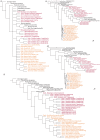
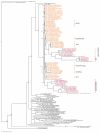
Results: The analysis of 110 Arsenophonus sequences (incl. Riesia and Phlomobacter), provides a robust monophyletic clade, characterized by unique molecular synapomorphies. On the other hand, unstable inner topology indicates that complete understanding of Arsenophonus evolution cannot be achieved with 16S rDNA. Moreover, taxonomically restricted Sampling matrices prove sensitivity of the phylogenetic signal to sampling; in some cases, Arsenophonus monophyly is disrupted by other symbiotic bacteria. Two contrasting coevolutionary patterns occur throughout the tree: parallel host-symbiont evolution and the haphazard association of the symbionts with distant hosts. A further conspicuous feature of the topology is the occurrence of monophyletic symbiont lineages associated with monophyletic groups of hosts without a co-speciation pattern. We suggest that part of this incongruence could be caused by methodological artifacts, such as intragenomic variability.
Conclusion: The sample of currently available molecular data presents the genus Arsenophonus as one of the richest and most widespread clusters of insect symbiotic bacteria. The analysis of its phylogenetic lineages indicates a complex evolution and apparent ecological versatility with switches between entirely different life styles. Due to these properties, the genus should play an important role in the studies of evolutionary trends in insect intracellular symbionts. However, under the current practice, relying exclusively on 16S rRNA sequences, the phylogenetic analyses are sensitive to various methodological artifacts that may even lead to description of new Arsenophonus lineages as independent genera (e.g. Riesia and Phlomobacter). The resolution of the evolutionary questions encountered within the Arsenophonus clade will thus require identification of new molecular markers suitable for the low-level phylogenetics.
Figures




Similar articles
-
Evidence of diversity and recombination in Arsenophonus symbionts of the Bemisia tabaci species complex.BMC Microbiol. 2012 Jan 18;12 Suppl 1(Suppl 1):S10. doi: 10.1186/1471-2180-12-S1-S10. BMC Microbiol. 2012. PMID: 22375811 Free PMC article.
-
An effect of 16S rRNA intercistronic variability on coevolutionary analysis in symbiotic bacteria: molecular phylogeny of Arsenophonus triatominarum.Syst Appl Microbiol. 2008 Jun;31(2):88-100. doi: 10.1016/j.syapm.2008.02.004. Epub 2008 May 16. Syst Appl Microbiol. 2008. PMID: 18485654
-
Multiple origins of endosymbiosis within the Enterobacteriaceae (γ-Proteobacteria): convergence of complex phylogenetic approaches.BMC Biol. 2011 Dec 28;9:87. doi: 10.1186/1741-7007-9-87. BMC Biol. 2011. PMID: 22201529 Free PMC article.
-
Evolution, multiple acquisition, and localization of endosymbionts in bat flies (Diptera: Hippoboscoidea: Streblidae and Nycteribiidae).Appl Environ Microbiol. 2013 May;79(9):2952-61. doi: 10.1128/AEM.03814-12. Epub 2013 Feb 22. Appl Environ Microbiol. 2013. PMID: 23435889 Free PMC article.
-
Winding paths to simplicity: genome evolution in facultative insect symbionts.FEMS Microbiol Rev. 2016 Nov 1;40(6):855-874. doi: 10.1093/femsre/fuw028. FEMS Microbiol Rev. 2016. PMID: 28204477 Free PMC article. Review.
Cited by
-
Characterization of the bacterial communities of psyllids associated with Rutaceae in Bhutan by high throughput sequencing.BMC Microbiol. 2020 Jul 20;20(1):215. doi: 10.1186/s12866-020-01895-4. BMC Microbiol. 2020. PMID: 32689950 Free PMC article.
-
Evidence of diversity and recombination in Arsenophonus symbionts of the Bemisia tabaci species complex.BMC Microbiol. 2012 Jan 18;12 Suppl 1(Suppl 1):S10. doi: 10.1186/1471-2180-12-S1-S10. BMC Microbiol. 2012. PMID: 22375811 Free PMC article.
-
Novel clade of alphaproteobacterial endosymbionts associated with stinkbugs and other arthropods.Appl Environ Microbiol. 2012 Jun;78(12):4149-56. doi: 10.1128/AEM.00673-12. Epub 2012 Apr 13. Appl Environ Microbiol. 2012. PMID: 22504806 Free PMC article.
-
Association between louse abundance and MHC II supertypes in Galápagos mockingbirds.Parasitol Res. 2020 May;119(5):1597-1605. doi: 10.1007/s00436-020-06617-3. Epub 2020 Jan 31. Parasitol Res. 2020. PMID: 32006226
-
Patterns of asexual reproduction of the soybean aphid, Aphis glycines (Matsumura), with and without the secondary symbionts Wolbachia and Arsenophonus, on susceptible and resistant soybean genotypes.Front Microbiol. 2023 Aug 31;14:1209595. doi: 10.3389/fmicb.2023.1209595. eCollection 2023. Front Microbiol. 2023. PMID: 37720159 Free PMC article.
References
-
- Gherna RL, Werren JH, Weisburg W, Cote R, Woese CR, Mandelco L, Brenner DJ. Arsenophonus nasoniae gen. nov., sp. nov., the causative agent of the son-killer trait in the parasitic wasp Nasonia vitripennis. Int J Syst Bacteriol. 1991;41:563–565.
-
- Hypša V. Endocytobionts of Triatoma infestans : distribution and transmission. J Invertebr Pathol. 1993;61:32–38. doi: 10.1006/jipa.1993.1006. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

