Zebrafish RNase T2 genes and the evolution of secretory ribonucleases in animals
- PMID: 19619322
- PMCID: PMC2720953
- DOI: 10.1186/1471-2148-9-170
Zebrafish RNase T2 genes and the evolution of secretory ribonucleases in animals
Abstract
Background: Members of the Ribonuclease (RNase) T2 family are common models for enzymological studies, and their evolution has been well characterized in plants. This family of acidic RNases is widespread, with members in almost all organisms including plants, animals, fungi, bacteria and even some viruses. While several biological functions have been proposed for these enzymes in plants, their role in animals is unknown. Interestingly, in vertebrates most of the biological roles of plant RNase T2 proteins are carried out by members of a different family, RNase A. Still, RNase T2 proteins are conserved in these animals
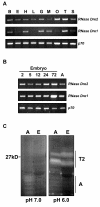
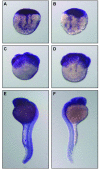
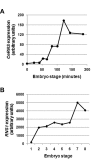
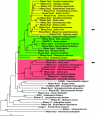
Results: As a first step to shed light on the role of animal RNase T2 enzymes, and to understand the evolution of these proteins while co-existing with the RNase A family, we characterized RNase Dre1 and RNase Dre2, the two RNase T2 genes present in the zebrafish (Danio rerio) genome. These genes are expressed in most tissues examined, including high expression in all stages of embryonic development, and their expression corresponds well with the presence of acidic RNase activities in every tissue analyzed. Embryo expression seems to be a conserved characteristic of members of this family, as other plant and animal RNase T2 genes show similar high expression during embryo development. While plant RNase T2 proteins and the vertebrate RNase A family show evidences of radiation and gene sorting, vertebrate RNase T2 proteins form a monophyletic group, but there is also another monophyletic group defining a fish-specific RNase T2 clade.
Conclusion: Based on gene expression and phylogenetic analyses we propose that RNase T2 enzymes carry out a housekeeping function. This conserved biological role probably kept RNase T2 enzymes in animal genomes in spite of the presence of RNases A. A hypothetical role during embryo development is also discussed.
Figures



Similar articles
-
RNase T2 genes from rice and the evolution of secretory ribonucleases in plants.Mol Genet Genomics. 2010 Apr;283(4):381-96. doi: 10.1007/s00438-010-0524-9. Epub 2010 Feb 25. Mol Genet Genomics. 2010. PMID: 20182746
-
Phylogenetic analyses and characterization of RNase X25 from Drosophila melanogaster suggest a conserved housekeeping role and additional functions for RNase T2 enzymes in protostomes.PLoS One. 2014 Aug 18;9(8):e105444. doi: 10.1371/journal.pone.0105444. eCollection 2014. PLoS One. 2014. PMID: 25133712 Free PMC article.
-
Ribonucleases with angiogenic and bactericidal activities from the Atlantic salmon.FEBS J. 2008 Mar;275(6):1283-95. doi: 10.1111/j.1742-4658.2008.06289.x. Epub 2008 Feb 12. FEBS J. 2008. PMID: 18279393
-
Ribonucleases from T2 family.Crit Rev Microbiol. 2002;28(2):79-122. doi: 10.1080/1040-840291046704. Crit Rev Microbiol. 2002. PMID: 12109772 Review.
-
The success of the RNase scaffold in the advance of biosciences and in evolution.Gene. 2007 Dec 30;406(1-2):8-12. doi: 10.1016/j.gene.2007.05.006. Epub 2007 May 21. Gene. 2007. PMID: 17616268 Review.
Cited by
-
Schistosome-derived omega-1 drives Th2 polarization by suppressing protein synthesis following internalization by the mannose receptor.J Exp Med. 2012 Sep 24;209(10):1753-67, S1. doi: 10.1084/jem.20111381. Epub 2012 Sep 10. J Exp Med. 2012. PMID: 22966004 Free PMC article.
-
RNS2, a conserved member of the RNase T2 family, is necessary for ribosomal RNA decay in plants.Proc Natl Acad Sci U S A. 2011 Jan 18;108(3):1093-8. doi: 10.1073/pnas.1009809108. Epub 2011 Jan 3. Proc Natl Acad Sci U S A. 2011. PMID: 21199950 Free PMC article.
-
Agrobacterium tumefaciens mediated transformation of the aquatic carnivorous plant Utricularia gibba.Plant Methods. 2020 Apr 10;16:50. doi: 10.1186/s13007-020-00592-7. eCollection 2020. Plant Methods. 2020. PMID: 32308728 Free PMC article.
-
Zebrafish disease model of human RNASET2-deficient cystic leukoencephalopathy displays abnormalities in early microglia.Biol Open. 2020 May 7;9(5):bio049239. doi: 10.1242/bio.049239. Biol Open. 2020. PMID: 32295832 Free PMC article.
-
Natural History of Innate Host Defense Peptides.Probiotics Antimicrob Proteins. 2009 Dec;1(2):97-112. doi: 10.1007/s12602-009-9031-x. Probiotics Antimicrob Proteins. 2009. PMID: 26783164
References
-
- Pizzo E, D'Alessio G. The success of the RNase scaffold in the advance of biosciences and in evolution. Gene. 2007;406:8–12. - PubMed
-
- Sato K, Egami F. Studies on ribonuclease in Takadiastase I. J Biochem. 1957;44:753–767.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

