Computer simulations of protein diffusion in compartmentalized cell membranes
- PMID: 19619461
- PMCID: PMC2711340
- DOI: 10.1016/j.bpj.2009.04.060
Computer simulations of protein diffusion in compartmentalized cell membranes
Abstract
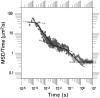
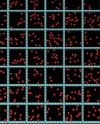
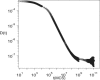
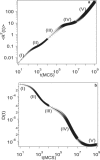
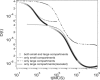
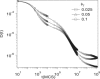
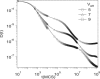
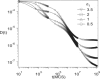
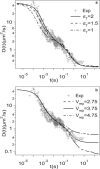
The diffusion of proteins in the cell membrane is investigated using computer simulations of a two-dimensional model. The membrane is assumed to be divided into compartments, with adjacent compartments separated by a barrier of stationary obstacles. Each compartment contains traps represented by stationary attractive disks. Depending on their size, these traps are intended to model either smaller compartments or binding sites. The simulations are intended to model the double-compartment model, which has been used to interpret single molecule experiments in normal rat kidney cells, where five regimes of transport are observed. The simulations show, however, that five regimes are observed only when there is a large separation between the sizes of the traps and large compartments, casting doubt on the double compartment model for the membrane. The diffusive behavior is sensitive to the concentration and size of traps and the strength of the barrier between compartments suggesting that the diffusion of proteins can be effectively used to characterize the structure of the membrane.
Figures
Similar articles
-
Monte Carlo investigation of diffusion of receptors and ligands that bind across opposing surfaces.Ann Biomed Eng. 2011 Jan;39(1):427-42. doi: 10.1007/s10439-010-0143-y. Epub 2010 Sep 2. Ann Biomed Eng. 2011. PMID: 20811955 Free PMC article.
-
Numerical calculation on a two-step subdiffusion behavior of lateral protein movement in plasma membranes.Phys Rev E. 2017 Oct;96(4-1):042410. doi: 10.1103/PhysRevE.96.042410. Epub 2017 Oct 19. Phys Rev E. 2017. PMID: 29347488
-
Sources of anomalous diffusion on cell membranes: a Monte Carlo study.Biophys J. 2007 Mar 15;92(6):1975-87. doi: 10.1529/biophysj.105.076869. Epub 2006 Dec 22. Biophys J. 2007. PMID: 17189312 Free PMC article.
-
Conditions for extreme sensitivity of protein diffusion in membranes to cell environments.Proc Natl Acad Sci U S A. 2006 Oct 10;103(41):15002-7. doi: 10.1073/pnas.0606992103. Epub 2006 Sep 28. Proc Natl Acad Sci U S A. 2006. PMID: 17008402 Free PMC article. Review.
-
Modeling 2D and 3D diffusion.Methods Mol Biol. 2007;400:295-321. doi: 10.1007/978-1-59745-519-0_20. Methods Mol Biol. 2007. PMID: 17951742 Review.
Cited by
-
Monte Carlo investigation of diffusion of receptors and ligands that bind across opposing surfaces.Ann Biomed Eng. 2011 Jan;39(1):427-42. doi: 10.1007/s10439-010-0143-y. Epub 2010 Sep 2. Ann Biomed Eng. 2011. PMID: 20811955 Free PMC article.
-
Lateral dynamics of proteins with polybasic domain on anionic membranes: a dynamic Monte-Carlo study.Biophys J. 2011 Mar 2;100(5):1261-70. doi: 10.1016/j.bpj.2011.01.025. Biophys J. 2011. PMID: 21354399 Free PMC article.
-
Membrane Compartmentalization Reducing the Mobility of Lipids and Proteins within a Model Plasma Membrane.J Phys Chem B. 2016 Sep 1;120(34):8873-81. doi: 10.1021/acs.jpcb.6b05846. Epub 2016 Aug 16. J Phys Chem B. 2016. PMID: 27483109 Free PMC article.
References
-
- Glaser M. Lipid domains in biological-membranes. Curr. Opin. Struct. Biol. 1993;3:475–481.
-
- Kusumi A., Nakada C., Ritchie K., Murase K., Suzuki K. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 2005;34:351–354. - PubMed
-
- Engelman D.M. Membranes are more mosaic than fluid. Nature. 2005;438:578–580. - PubMed
-
- Saxton M.J., Jacobson K. Single-particle tracking: applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 1997;26:373–399. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

