A novel gene essential for the development of single positive thymocytes
- PMID: 19620281
- PMCID: PMC2738282
- DOI: 10.1128/MCB.00793-09
A novel gene essential for the development of single positive thymocytes
Abstract
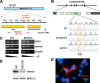
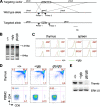
A critical step during intrathymic T-cell development is the transition of CD4(+) CD8(+) double-positive (DP) cells to the major histocompatibility complex class I (MHC-I)-restricted CD4(-) CD8(+) and MHC-II-restricted CD4(+) CD8(-) single-positive (SP) cell stage. Here, we identify a novel gene that is essential for this process. Through the T-cell phenotype-based screening of N-ethyl-N-nitrosourea (ENU)-induced mutant mice, we established a mouse line in which numbers of CD4 and CD8 SP thymocytes as well as peripheral CD4 and CD8 T cells were dramatically reduced. Using linkage analysis and DNA sequencing, we identified a missense point mutation in a gene, E430004N04Rik (also known as themis), that does not belong to any known gene family. This orphan gene is expressed specifically in DP and SP thymocytes and peripheral T cells, whereas in mutant thymocytes the levels of protein encoded by this gene were drastically reduced. We generated E430004N04Rik-deficient mice, and their phenotype was virtually identical to that of the ENU mutant mice, thereby confirming that this gene is essential for the development of SP thymocytes.
Figures




Similar articles
-
A novel mouse thymocyte antigen (F3Ag): down-regulation during the CD4+CD8+ double-positive stage indicates positive selection.Int Immunol. 1996 Jan;8(1):101-13. doi: 10.1093/intimm/8.1.101. Int Immunol. 1996. PMID: 8671594
-
Runx3 regulates integrin alpha E/CD103 and CD4 expression during development of CD4-/CD8+ T cells.J Immunol. 2005 Aug 1;175(3):1694-705. doi: 10.4049/jimmunol.175.3.1694. J Immunol. 2005. PMID: 16034110
-
Analyzing expression of perforin, Runx3, and Thpok genes during positive selection reveals activation of CD8-differentiation programs by MHC II-signaled thymocytes.J Immunol. 2005 Oct 1;175(7):4465-74. doi: 10.4049/jimmunol.175.7.4465. J Immunol. 2005. PMID: 16177089
-
Fc gamma RII/III and CD2 expression mark distinct subpopulations of immature CD4-CD8- murine thymocytes: in vivo developmental kinetics and T cell receptor beta chain rearrangement status.J Exp Med. 1993 Apr 1;177(4):1079-92. doi: 10.1084/jem.177.4.1079. J Exp Med. 1993. PMID: 8096236 Free PMC article.
-
The final maturation of at least some single-positive CD4(hi) thymocytes does not require T cell receptor-major histocompatibility complex contact.J Exp Med. 1999 Sep 20;190(6):757-64. doi: 10.1084/jem.190.6.757. J Exp Med. 1999. PMID: 10499914 Free PMC article.
Cited by
-
T cell receptor signaling for γδT cell development.Inflamm Regen. 2019 Mar 28;39:6. doi: 10.1186/s41232-019-0095-z. eCollection 2019. Inflamm Regen. 2019. PMID: 30976362 Free PMC article. Review.
-
THEMIS increases TCR signaling in CD4+CD8+ thymocytes by inhibiting the activity of the tyrosine phosphatase SHP1.Sci Signal. 2023 May 9;16(784):eade1274. doi: 10.1126/scisignal.ade1274. Epub 2023 May 9. Sci Signal. 2023. PMID: 37159521 Free PMC article.
-
Themis controls T cell activation, effector functions, and metabolism of peripheral CD8+ T cells.Life Sci Alliance. 2023 Sep 22;6(12):e202302156. doi: 10.26508/lsa.202302156. Print 2023 Dec. Life Sci Alliance. 2023. PMID: 37739454 Free PMC article.
-
Themis suppresses the effector function of CD8+ T cells in acute viral infection.Cell Mol Immunol. 2023 May;20(5):512-524. doi: 10.1038/s41423-023-00997-z. Epub 2023 Mar 28. Cell Mol Immunol. 2023. PMID: 36977779 Free PMC article.
-
A THEMIS:SHP1 complex promotes T-cell survival.EMBO J. 2015 Feb 3;34(3):393-409. doi: 10.15252/embj.201387725. Epub 2014 Dec 22. EMBO J. 2015. PMID: 25535246 Free PMC article.
References
-
- Asano, A., K. Tsubomatsu, C. G. Jung, N. Sasaki, and T. Agui. 2007. A deletion mutation of the protein tyrosine phosphatase kappa (Ptprk) gene is responsible for T-helper immunodeficiency (thid) in the LEC rat. Mamm. Genome 18779-786. - PubMed
-
- Benveniste, P., G. Knowles, and A. Cohen. 1996. CD8/CD4 lineage commitment occurs by an instructional/default process followed by positive selection. Eur. J. Immunol. 26461-471. - PubMed
-
- Cook, M. C., C. G. Vinuesa, and C. C. Goodnow. 2006. ENU-mutagenesis: insight into immune function and pathology. Curr. Opin. Immunol. 18627-633. - PubMed
-
- Germain, R. N. 2002. T-cell development and the CD4-CD8 lineage decision. Nat. Rev. Immunol. 2309-322. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
