The fission yeast HIRA histone chaperone is required for promoter silencing and the suppression of cryptic antisense transcripts
- PMID: 19620282
- PMCID: PMC2738286
- DOI: 10.1128/MCB.00698-09
The fission yeast HIRA histone chaperone is required for promoter silencing and the suppression of cryptic antisense transcripts
Abstract
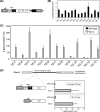
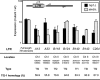
The assembly of nucleosomes by histone chaperones is an important component of transcriptional regulation. Here, we have assessed the global roles of the HIRA histone chaperone in Schizosaccharomyces pombe. Microarray analysis indicates that inactivation of the HIRA complex results in increased expression of at least 4% of fission yeast genes. HIRA-regulated genes overlap with those which are normally repressed in vegetatively growing cells, such as targets of the Clr6 histone deacetylase and silenced genes located in subtelomeric regions. HIRA is also required for silencing of all 13 intact copies of the Tf2 long terminal repeat (LTR) retrotransposon. However, the role of HIRA is not restricted to bona fide promoters, because HIRA also suppresses noncoding transcripts from solo LTR elements and spurious antisense transcripts from cryptic promoters associated with transcribed regions. Furthermore, the HIRA complex is essential in the absence of the quality control provided by nuclear exosome-mediated degradation of illegitimate transcripts. This suggests that HIRA restricts genomic accessibility, and consistent with this, the chromosomes of cells lacking HIRA are more susceptible to genotoxic agents that cause double-strand breaks. Thus, the HIRA histone chaperone is required to maintain the protective functions of chromatin.
Figures


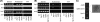
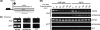
References
-
- Ahmad, A., Y. Takami, and T. Nakayama. 2004. WD dipeptide motifs and LXXLL motif of chicken HIRA are essential for interactions with the p48 subunit of chromatin assembly factor-1 and histone deacetylase-2 in vitro and in vivo. Gene 342125-136. - PubMed
-
- Ahmad, A., Y. Takami, and T. Nakayama. 2003. WD dipeptide motifs and LXXLL motif of chicken HIRA are necessary for transcription repression and the latter motif is essential for interaction with histone deacetylase-2 in vivo. Biochem. Biophys. Res. Commun. 3121266-1272. - PubMed
-
- Ahmad, K., and S. Henikoff. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 91191-1200. - PubMed
-
- Blackwell, C., K. A. Martin, A. Greenall, A. Pidoux, R. C. Allshire, and S. K. Whitehall. 2004. The Schizosaccharomyces pombe HIRA-like protein Hip1 is required for the periodic expression of histone genes and contributes to the function of complex centromeres. Mol. Cell. Biol. 244309-4320. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
