The CENP-S complex is essential for the stable assembly of outer kinetochore structure
- PMID: 19620631
- PMCID: PMC2717651
- DOI: 10.1083/jcb.200903100
The CENP-S complex is essential for the stable assembly of outer kinetochore structure
Abstract
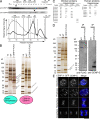
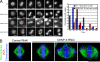
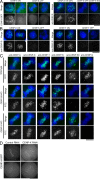
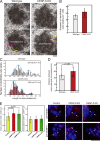
The constitutive centromere-associated network (CCAN) proteins are central to kinetochore assembly. To define the molecular architecture of this critical kinetochore network, we sought to determine the full complement of CCAN components and to define their relationships. This work identified a centromere protein S (CENP-S)-containing subcomplex that includes the new constitutive kinetochore protein CENP-X. Both CENP-S- and CENP-X-deficient chicken DT40 cells are viable but show abnormal mitotic behavior based on live cell analysis. Human HeLa cells depleted for CENP-X also showed mitotic errors. The kinetochore localization of CENP-S and -X is abolished in CENP-T- or CENP-K-deficient cells, but reciprocal experiments using CENP-S-deficient cells did not reveal defects in the localization of CCAN components. However, CENP-S- and CENP-X-deficient cells show a significant reduction in the size of the kinetochore outer plate. In addition, we found that intrakinetochore distance was increased in CENP-S- and CENP-X-deficient cells. These results suggest that the CENP-S complex is essential for the stable assembly of the outer kinetochore.
Figures
References
-
- Cheeseman I.M., Desai A.. 2008. Molecular architecture of the kinetochore-microtubule interface.Nat. Rev. Mol. Cell Biol. 9:33–46. - PubMed
-
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., Desai A.. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore.Cell. 127:983–997. - PubMed
-
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., Salmon E.D.. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1.Cell. 127:969–982. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

