The JNKs differentially regulate RNA polymerase III transcription by coordinately modulating the expression of all TFIIIB subunits
- PMID: 19620725
- PMCID: PMC2722300
- DOI: 10.1073/pnas.0904843106
The JNKs differentially regulate RNA polymerase III transcription by coordinately modulating the expression of all TFIIIB subunits
Abstract
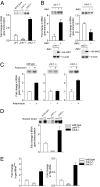
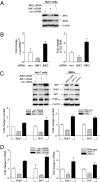
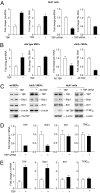
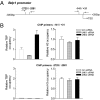
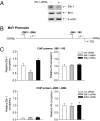
RNA polymerase (pol) III-dependent transcription is subject to stringent regulation by tumor suppressors and oncogenic proteins and enhanced RNA pol III transcription is essential for cellular transformation and tumorigenesis. Since the c-Jun N-terminal kinases (JNKs) display both oncogenic and tumor suppressor properties, the roles of these proteins in regulating RNA pol III transcription were examined. In both mouse and human cells, loss or reduction in JNK1 expression represses RNA pol III transcription. In contrast, loss or reduction in JNK2 expression induces transcription. The JNKs coordinately regulate expression of all 3 TFIIIB subunits. While JNK1 positively regulates TBP expression, the RNA pol III-specific factors, Brf1 and Bdp1, JNK2 negatively regulates their expression. Brf1 is coregulated with TBP through the JNK target, Elk-1. Reducing Elk-1 expression decreases Brf1 expression. Decreasing JNK1 expression reduces Elk-1 occupancy at the Brf1 promoter, while decreasing JNK2 expression enhances recruitment of Elk-1 to the Brf1 promoter. In contrast, regulation of Bdp1 occurs through JNK-mediated alterations in TBP expression. Altered TBP expression mimics the effect of reduced JNK1 or JNK2 levels on Bdp1 expression. Decreasing JNK1 expression reduces the occupancy of TBP at the Bdp1 promoter, while decreasing JNK2 expression enhances recruitment of TBP to the Bdp1 promoter. Together, these results provide a molecular mechanism for regulating RNA pol III transcription through the coordinate control of TFIIIB subunit expression and elucidate opposing functions for the JNKs in regulating a large class of genes that dictate the biosynthetic capacity of cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. - PubMed
-
- Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. - PubMed
-
- Ventura JJ, Hübner A, Zhang C, Flavell RA, Shokat KM, Davis RJ. Chemical genetic analysis of the time course of signal transduction by JNK. Mol Cell. 2006;5:701–710. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

