Nucleotide-dependent conformational states of actin
- PMID: 19620726
- PMCID: PMC2722336
- DOI: 10.1073/pnas.0902092106
Nucleotide-dependent conformational states of actin
Abstract
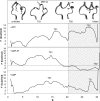
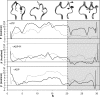
The influence of the state of the bound nucleotide (ATP, ADP-Pi, or ADP) on the conformational free-energy landscape of actin is investigated. Nucleotide-dependent folding of the DNase-I binding (DB) loop in monomeric actin and the actin trimer is carried out using all-atom molecular dynamics (MD) calculations accelerated with a multiscale implementation of the metadynamics algorithm. Additionally, an investigation of the opening and closing of the actin nucleotide binding cleft is performed. Nucleotide-dependent free-energy profiles for all of these conformational changes are calculated within the framework of metadynamics. We find that in ADP-bound monomer, the folded and unfolded states of the DB loop have similar relative free-energy. This result helps explain the experimental difficulty in obtaining an ordered crystal structure for this region of monomeric actin. However, we find that in the ADP-bound actin trimer, the folded DB loop is stable and in a free-energy minimum. It is also demonstrated that the nucleotide binding cleft favors a closed conformation for the bound nucleotide in the ATP and ADP-Pi states, whereas the ADP state favors an open confirmation, both in the monomer and trimer. These results suggest a mechanism of allosteric interactions between the nucleotide binding cleft and the DB loop. This behavior is confirmed by an additional simulation that shows the folding free-energy as a function of the nucleotide cleft width, which demonstrates that the barrier for folding changes significantly depending on the value of the cleft width.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
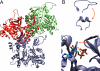

Similar articles
-
Nucleotide effects on the structure and dynamics of actin.Biophys J. 2007 Aug 15;93(4):1277-83. doi: 10.1529/biophysj.107.109215. Epub 2007 May 25. Biophys J. 2007. PMID: 17526584 Free PMC article.
-
Allostery of actin filaments: molecular dynamics simulations and coarse-grained analysis.Proc Natl Acad Sci U S A. 2005 Sep 13;102(37):13111-6. doi: 10.1073/pnas.0503732102. Epub 2005 Aug 31. Proc Natl Acad Sci U S A. 2005. PMID: 16135566 Free PMC article.
-
The crystal structure of uncomplexed actin in the ADP state.Science. 2001 Jul 27;293(5530):708-11. doi: 10.1126/science.1059700. Science. 2001. PMID: 11474115
-
Actin polymerization: regulation by divalent metal ion and nucleotide binding, ATP hydrolysis and binding of myosin.Adv Exp Med Biol. 1994;358:71-81. doi: 10.1007/978-1-4615-2578-3_7. Adv Exp Med Biol. 1994. PMID: 7801813 Review.
-
ATPase activity and conformational changes in the regulation of actin.Biochim Biophys Acta. 2001 Oct 18;1549(2):137-47. doi: 10.1016/s0167-4838(01)00255-2. Biochim Biophys Acta. 2001. PMID: 11690650 Review.
Cited by
-
The allosteric switching mechanism in bacteriophage MS2.J Chem Phys. 2016 Jul 21;145(3):035101. doi: 10.1063/1.4955187. J Chem Phys. 2016. PMID: 27448905 Free PMC article.
-
Actin R256 Mono-methylation Is a Conserved Post-translational Modification Involved in Transcription.Cell Rep. 2020 Sep 29;32(13):108172. doi: 10.1016/j.celrep.2020.108172. Cell Rep. 2020. PMID: 32997990 Free PMC article.
-
Defining coarse-grained representations of large biomolecules and biomolecular complexes from elastic network models.Biophys J. 2009 Oct 21;97(8):2327-37. doi: 10.1016/j.bpj.2009.08.007. Biophys J. 2009. PMID: 19843465 Free PMC article.
-
Mutation of actin Tyr-53 alters the conformations of the DNase I-binding loop and the nucleotide-binding cleft.J Biol Chem. 2010 Mar 26;285(13):9729-9739. doi: 10.1074/jbc.M109.073452. Epub 2010 Jan 25. J Biol Chem. 2010. PMID: 20100837 Free PMC article.
-
Nucleotide regulation of the structure and dynamics of G-actin.Biophys J. 2014 Apr 15;106(8):1710-20. doi: 10.1016/j.bpj.2014.03.012. Biophys J. 2014. PMID: 24739170 Free PMC article.
References
-
- Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. - PubMed
-
- Reisler E, Egelman EH. Actin's structure and function: What we still do not understand. J Biol Chem. 2007;282:36133–36137. - PubMed
-
- Carlier MF, Pantaloni D. Direct evidence for ADP-Pi-F-actin as the major intermediate in ATP-actin polymerization. Rate of dissociation of Pi from actin-filaments. Biochemistry. 1986;25:7789–7792. - PubMed
-
- Isambert H, et al. Flexibility of actin filaments derived from thermal fluctuations. Effect of bound nucleotide, phalloidin, and muscle regulatory proteins. J Biol Chem. 1995;270:11437–11444. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

