Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo
- PMID: 19620965
- PMCID: PMC2823114
- DOI: 10.1038/nsmb.1636
Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo
Abstract
We assess the role of intrinsic histone-DNA interactions by mapping nucleosomes assembled in vitro on genomic DNA. Nucleosomes strongly prefer yeast DNA over Escherichia coli DNA, indicating that the yeast genome evolved to favor nucleosome formation. Many yeast promoter and terminator regions intrinsically disfavor nucleosome formation, and nucleosomes assembled in vitro show strong rotational positioning. Nucleosome arrays generated by the ACF assembly factor have fewer nucleosome-free regions, reduced rotational positioning and less translational positioning than obtained by intrinsic histone-DNA interactions. Notably, nucleosomes assembled in vitro have only a limited preference for specific translational positions and do not show the pattern observed in vivo. Our results argue against a genomic code for nucleosome positioning, and they suggest that the nucleosomal pattern in coding regions arises primarily from statistical positioning from a barrier near the promoter that involves some aspect of transcriptional initiation by RNA polymerase II.
Figures
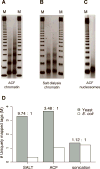
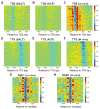
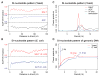

Comment in
-
Nucleosome sequence preferences influence in vivo nucleosome organization.Nat Struct Mol Biol. 2010 Aug;17(8):918-20. doi: 10.1038/nsmb0810-918. Nat Struct Mol Biol. 2010. PMID: 20683473 Free PMC article. No abstract available.
References
-
- Kornberg RD. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. - PubMed
-
- Yuan GC, Liu YJ, Dion MF, et al. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. - PubMed
-
- Lee W, Tillo D, Bray N, et al. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–1244. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

