Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry
- PMID: 19620975
- PMCID: PMC3203213
- DOI: 10.1038/nn.2371
Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry
Abstract
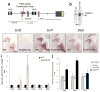
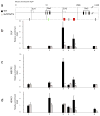
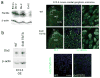
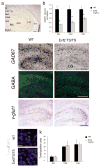
Genomic studies demonstrate that, although the majority of the mammalian genome is transcribed, only about 2% of these transcripts are code for proteins. We investigated how the long, polyadenylated Evf2 noncoding RNA regulates transcription of the homeodomain transcription factors DLX5 and DLX6 in the developing mouse forebrain. We found that, in developing ventral forebrain, Evf2 recruited DLX and MECP2 transcription factors to important DNA regulatory elements in the Dlx5/6 intergenic region and controlled Dlx5, Dlx6 and Gad1 expression through trans and cis-acting mechanisms. Evf2 mouse mutants had reduced numbers of GABAergic interneurons in early postnatal hippocampus and dentate gyrus. Although the numbers of GABAergic interneurons and Gad1 RNA levels returned to normal in Evf2 mutant adult hippocampus, reduced synaptic inhibition occurred. These results suggest that noncoding RNA-dependent balanced gene regulation in embryonic brain is critical for proper formation of GABA-dependent neuronal circuitry in adult brain.
Figures
References
-
- Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21:11–42. - PubMed
-
- Mattick JS. A new paradigm for developmental biology. J Exp Biol. 2007;210:1526–1547. - PubMed
-
- Shamovsky I, Nudler E. Gene control by large noncoding RNAs. Sci STKE. 2006;2006:pe40. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

