Enzymatic transition states and dynamic motion in barrier crossing
- PMID: 19620996
- PMCID: PMC2859820
- DOI: 10.1038/nchembio.202
Enzymatic transition states and dynamic motion in barrier crossing
Abstract
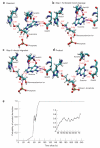
What are the atomic motions at enzymatic catalytic sites on the timescale of chemical change? Combined experimental and computational chemistry approaches take advantage of transition-state analogs to reveal dynamic motions linked to transition-state formation. QM/MM transition path sampling from reactive complexes provides both temporal and dynamic information for barrier crossing. Fast (femtosecond to picosecond) dynamic motions provide essential links to enzymatic barrier crossing by local or promoting-mode dynamic searches through bond-vibrational space. Transition-state lifetimes are within the femtosecond timescales of bond vibrations and show no manifestations of stabilized, equilibrated complexes. The slow binding and protein conformational changes (microsecond to millisecond) also required for catalysis are temporally decoupled from the fast dynamic motions forming the transition state. According to this view of enzymatic catalysis, transition states are formed by fast, coincident dynamic excursions of catalytic site elements, while the binding of transition-state analogs is the conversion of the dynamic excursions to equilibrated states.
Figures





References
-
- Pauling LC. Molecular architecture and biological reactions. Chem. Eng. News. 1946;24:1375–1377.
-
- Wolfenden R. Transition state analogues for enzyme catalysis. Nature. 1969;223:704–705. - PubMed
-
- Hammes-Schiffer S, Benkovic SJ. Relating protein motion to catalysis. Annu. Rev. Biochem. 2006;75:519–541. - PubMed
-
- Schramm VL. Enzymatic transition state theory and transition state analogue design. J. Biol. Chem. 2007;282:28297–28300. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information

