Novel transthyretin amyloid fibril formation inhibitors: synthesis, biological evaluation, and X-ray structural analysis
- PMID: 19621084
- PMCID: PMC2709434
- DOI: 10.1371/journal.pone.0006290
Novel transthyretin amyloid fibril formation inhibitors: synthesis, biological evaluation, and X-ray structural analysis
Abstract
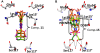
Transthyretin (TTR) is one of thirty non-homologous proteins whose misfolding, dissociation, aggregation, and deposition is linked to human amyloid diseases. Previous studies have identified that TTR amyloidogenesis can be inhibited through stabilization of the native tetramer state by small molecule binding to the thyroid hormone sites of TTR. We have evaluated a new series of beta-aminoxypropionic acids (compounds 5-21), with a single aromatic moiety (aryl or fluorenyl) linked through a flexible oxime tether to a carboxylic acid. These compounds are structurally distinct from the native ligand thyroxine and typical halogenated biaryl NSAID-like inhibitors to avoid off-target hormonal or anti-inflammatory activity. Based on an in vitro fibril formation assay, five of these compounds showed significant inhibition of TTR amyloidogenesis, with two fluorenyl compounds displaying inhibitor efficacy comparable to the well-known TTR inhibitor diflunisal. Fluorenyl 15 is the most potent compound in this series and importantly does not show off-target anti-inflammatory activity. Crystal structures of the TTR:inhibitor complexes, in agreement with molecular docking studies, revealed that the aromatic moiety, linked to the sp(2)-hybridized oxime carbon, specifically directed the ligand in either a forward or reverse binding mode. Compared to the aryl family members, the bulkier fluorenyl analogs achieved more extensive interactions with the binding pockets of TTR and demonstrated better inhibitory activity in the fibril formation assay. Preliminary optimization efforts are described that focused on replacement of the C-terminal acid in both the aryl and fluorenyl series (compounds 22-32). The compounds presented here constitute a new class of TTR inhibitors that may hold promise in treating amyloid diseases associated with TTR misfolding.
Conflict of interest statement
Figures
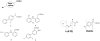
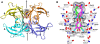

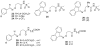

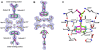

References
-
- Blake CC, Geisow MJ, Oatley SJ, Rerat B, Rerat C. Structure of prealbumin: secondary, tertiary and quaternary interactions determined by Fourier refinement at 1.8 A. J Mol Biol. 1978;121:339–356. - PubMed
-
- Richardson SJ, Bradley AJ, Duan W, Wettenhall RE, Harms PJ, et al. Evolution of marsupial and other vertebrate thyroxine-binding plasma proteins. Am J Physiol. 1994;266:R1359–1370. - PubMed
-
- Nilsson SF, Rask L, Peterson PA. Studies on thyroid hormone-binding proteins. II. Binding of thyroid hormones, retinol-binding protein, and fluorescent probes to prealbumin and effects of thyroxine on prealbumin subunit self association. J Biol Chem. 1975;250:8554–8563. - PubMed
-
- Wojtczak A, Cody V, Luft JR, Pangborn W. Structures of human transthyretin complexed with thyroxine at 2.0 A resolution and 3′,5′-dinitro-N-acetyl-L-thyronine at 2.2 A resolution. Acta Crystallogr D Biol Crystallogr. 1996;52:758–765. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

