Physical basis of metal-binding specificity in Escherichia coli NikR
- PMID: 19621966
- PMCID: PMC3579654
- DOI: 10.1021/ja9026314
Physical basis of metal-binding specificity in Escherichia coli NikR
Abstract
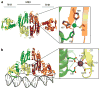
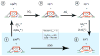
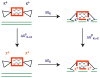
In Escherichia coli and other bacteria, nickel uptake is regulated by the transcription factor NikR. Nickel binding at high-affinity sites in E. coli NikR (EcNikR) facilitates EcNikR binding to the nik operon, where it then suppresses transcription of genes encoding the nickel uptake transporter, NikABCDE. A structure of the EcNikR-DNA complex suggests that a second metal-binding site is also present when NikR binds to the nik operon. Moreover, this co-crystal structure raises the question of what metal occupies the second site under physiological conditions: K(+), which is present in the crystal structure, or Ni(2+), which has been proposed to bind to low- as well as high-affinity sites on EcNikR. To determine which ion is preferred at the second metal-binding site and the physical basis for any preference of one ion over another in both the second metal-binding site and the high-affinity sites, we conducted a series of detailed molecular simulations on the EcNikR structure. Simulations that place Ni(2+) at high-affinity sites lead to stable trajectories with realistic ion-ligand distances and geometries, while simulations that place K(+) at these sites lead to conformational changes in the protein that are likely unfavorable for ion binding. By contrast, simulations on the second metal site in the EcNikR-DNA complex lead to stable trajectories with realistic geometries regardless of whether K(+) or Ni(2+) occupies this site. Electrostatic binding free energy calculations, however, suggest that EcNikR binding to DNA is more favorable when the second metal-binding site contains K(+). An analysis of the energetic contributions to the electrostatic binding free energy suggests that, while the interaction between EcNikR and DNA is more favorable when the second site contains Ni(2+), the large desolvation penalty associated with moving Ni(2+) from solution to the relatively buried second site offsets this favorable interaction term. Additional free energy simulations that account for both electrostatic and non-electrostatic effects argue that EcNikR binding to DNA is most favorable when the second site contains a monovalent ion the size of K(+). Taken together, these data suggest that the EcNikR structure is most stable when Ni(2+) occupies high-affinity sites and that EcNikR binding to DNA is more favorable when the second site contains K(+).
Figures

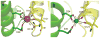


Similar articles
-
Nickel-specific response in the transcriptional regulator, Escherichia coli NikR.J Am Chem Soc. 2007 Apr 25;129(16):5085-95. doi: 10.1021/ja068505y. Epub 2007 Mar 31. J Am Chem Soc. 2007. PMID: 17397155
-
Metal-selective DNA-binding response of Escherichia coli NikR.Biochemistry. 2004 Aug 10;43(31):10029-38. doi: 10.1021/bi049404k. Biochemistry. 2004. PMID: 15287730
-
pH dependent Ni(II) binding and aggregation of Escherichia coli and Helicobacter pylori NikR.Biochimie. 2006 Nov;88(11):1693-705. doi: 10.1016/j.biochi.2006.07.016. Epub 2006 Aug 14. Biochimie. 2006. PMID: 16930800
-
Coordinating intracellular nickel-metal-site structure-function relationships and the NikR and RcnR repressors.Nat Prod Rep. 2010 May;27(5):658-67. doi: 10.1039/b906683g. Epub 2010 Mar 5. Nat Prod Rep. 2010. PMID: 20442957 Review.
-
Nickel transport systems in microorganisms.Arch Microbiol. 2000 Jan;173(1):1-9. doi: 10.1007/s002030050001. Arch Microbiol. 2000. PMID: 10648098 Review.
Cited by
-
Structural basis of low-affinity nickel binding to the nickel-responsive transcription factor NikR from Escherichia coli.Biochemistry. 2010 Sep 14;49(36):7830-8. doi: 10.1021/bi100923j. Biochemistry. 2010. PMID: 20704276 Free PMC article.
-
Metal site occupancy and allosteric switching in bacterial metal sensor proteins.Arch Biochem Biophys. 2012 Mar 15;519(2):210-22. doi: 10.1016/j.abb.2011.11.021. Epub 2011 Dec 8. Arch Biochem Biophys. 2012. PMID: 22178748 Free PMC article. Review.
-
Structural and mechanistic insights into Helicobacter pylori NikR activation.Nucleic Acids Res. 2010 May;38(9):3106-18. doi: 10.1093/nar/gkp1216. Epub 2010 Jan 19. Nucleic Acids Res. 2010. PMID: 20089510 Free PMC article.
-
Apo and nickel-bound forms of the Pyrococcus horikoshii species of the metalloregulatory protein: NikR characterized by molecular dynamics simulations.Biochemistry. 2009 Dec 22;48(50):12024-33. doi: 10.1021/bi9013352. Biochemistry. 2009. PMID: 19891498 Free PMC article.
-
Molecular Mechanism for Attractant Signaling to DHMA by E. coli Tsr.Biophys J. 2020 Jan 21;118(2):492-504. doi: 10.1016/j.bpj.2019.11.3382. Epub 2019 Nov 27. Biophys J. 2020. PMID: 31839263 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

