Divergent evolution in the cytoplasmic domains of PRLR and GHR genes in Artiodactyla
- PMID: 19622175
- PMCID: PMC2720954
- DOI: 10.1186/1471-2148-9-172
Divergent evolution in the cytoplasmic domains of PRLR and GHR genes in Artiodactyla
Abstract
Background: Prolactin receptor (PRLR) and growth hormone receptor (GHR) belong to the large superfamily of class 1 cytokine receptors. Both of them have been identified as candidate genes affecting key quantitative traits, like growth and reproduction in livestock. We have previously studied the molecular anatomy of the cytoplasmic domain of GHR in different cattle breeds and artiodactyl species. In this study we have analysed the corresponding cytoplasmic signalling region of PRLR.
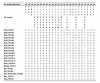
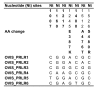
Results: We sequenced PRLR gene exon 10, coding for the major part of the cytoplasmic domain, from cattle, American bison, European bison, yak, sheep, pig and wild boar individuals. We found different patterns of variation in the two receptors within and between ruminants and pigs. Pigs and bison species have no variation within GHR exon 10, but show high haplotype diversity for the PRLR exon 10. In cattle, PRLR shows lower diversity than GHR. The Bovinae PRLR haplotype network fits better the known phylogenetic relationships between the species than that of the GHR, where differences within cattle breeds are larger than between the different species in the subfamily. By comparison with the wild boar haplotypes, a high number of subsequent nonsynonymous substitutions seem to have accumulated in the pig PRLR exon 10 after domestication.
Conclusion: Both genes affect a multitude of traits that have been targets of selection after domestication. The genes seem to have responded differently to different selection pressures imposed by human artificial selection. The results suggest possible effects of selective sweeps in GHR before domestication in the pig lineage or species divergence in the Bison lineage. The PRLR results may be explained by strong directional selection in pigs or functional switching.
Figures

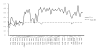

Similar articles
-
The role of the bovine growth hormone receptor and prolactin receptor genes in milk, fat and protein production in Finnish Ayrshire dairy cattle.Genetics. 2006 Aug;173(4):2151-64. doi: 10.1534/genetics.105.046730. Epub 2006 Jun 4. Genetics. 2006. PMID: 16751675 Free PMC article.
-
Molecular anatomy of the cytoplasmic domain of bovine growth hormone receptor, a quantitative trait locus.Proc Biol Sci. 2008 Jul 7;275(1642):1525-34. doi: 10.1098/rspb.2008.0181. Proc Biol Sci. 2008. PMID: 18381258 Free PMC article.
-
Molecular evolution of coding and non-coding sequences of the growth hormone receptor (GHR) gene in the family Bovidae.Folia Biol (Krakow). 2006;54(1-2):31-6. doi: 10.3409/173491606777919148. Folia Biol (Krakow). 2006. PMID: 17044257
-
Growth hormone (GH) binding and effects of GH analogs in transgenic mice.Proc Soc Exp Biol Med. 1994 Jul;206(3):190-4. doi: 10.3181/00379727-206-43740. Proc Soc Exp Biol Med. 1994. PMID: 8016152 Review.
-
The role of prolactin and growth hormone in mammary gland development.Mol Cell Endocrinol. 2002 Nov 29;197(1-2):127-31. doi: 10.1016/s0303-7207(02)00286-1. Mol Cell Endocrinol. 2002. PMID: 12431805 Review.
Cited by
-
A microsatellite-based analysis for the detection of selection on BTA1 and BTA20 in northern Eurasian cattle (Bos taurus) populations.Genet Sel Evol. 2010 Aug 6;42(1):32. doi: 10.1186/1297-9686-42-32. Genet Sel Evol. 2010. PMID: 20691068 Free PMC article.
-
European wild boars and domestic pigs display different polymorphic patterns in the Toll-like receptor (TLR) 1, TLR2, and TLR6 genes.Immunogenetics. 2010 Jan;62(1):49-58. doi: 10.1007/s00251-009-0409-4. Epub 2009 Dec 2. Immunogenetics. 2010. PMID: 19953243
-
Polymorphisms of the PRLR Gene and Their Association with Milk Production Traits in Egyptian Buffaloes.Animals (Basel). 2021 Apr 25;11(5):1237. doi: 10.3390/ani11051237. Animals (Basel). 2021. PMID: 33923003 Free PMC article.
-
Single nucleotide polymorphisms of the prolactin receptor (PRLR) gene and its association with growth traits in Chinese cattle.Mol Biol Rep. 2011 Jan;38(1):261-6. doi: 10.1007/s11033-010-0103-5. Epub 2010 Mar 27. Mol Biol Rep. 2011. PMID: 20349144
-
Convergent Evolution of Slick Coat in Cattle through Truncation Mutations in the Prolactin Receptor.Front Genet. 2018 Feb 23;9:57. doi: 10.3389/fgene.2018.00057. eCollection 2018. Front Genet. 2018. PMID: 29527221 Free PMC article.
References
-
- Evershed RP, Payne S, Sherratt AG, Copley MS, Coolidge J, Urem-Kotsu D, Kotsakis K, Ozdogan M, Ozdogan AE, Nieuwenhuyse O, Akkermans PM, Bailey D, Andeescu RR, Campbell S, Farid S, Hodder I, Yalman N, Ozbasaran M, Bicakci E, Garfinkel Y, Levy T, Burton MM. Earliest date for milk use in the Near East and southeastern Europe linked to cattle herding. Nature. 2008;455:528–531. doi: 10.1038/nature07180. - DOI - PubMed
-
- Bovine HapMap Consortium. Gibbs RA, Taylor JF, Van Tassell CP, Barendse W, Eversole KA, Gill CA, Green RD, Hamernik DL, Kappes SM, Lien S, Matukumalli LK, McEwan JC, Nazareth LV, Schnabel RD, Weinstock GM, Wheeler DA, Ajmone-Marsan P, Boettcher PJ, Caetano AR, Garcia JF, Hanotte O, Mariani P, Skow LC, Sonstegard TS, Williams JL, Diallo B, Hailemariam L, Martinez ML, Morris CA, Silva LO, Spelman RJ, Mulatu W, Zhao K, Abbey CA, Agaba M, Araujo FR, Bunch RJ, Burton J, Gorni C, Olivier H, Harrison BE, Luff B, Machado MA, Mwakaya J, Plastow G, Sim W, Smith T, Thomas MB, Valentini A, Williams P, Womack J, Woolliams JA, Liu Y, Qin X, Worley KC, Gao C, Jiang H, Moore SS, Ren Y, Song XZ, Bustamante CD, Hernandez RD, Muzny DM, Patil S, San Lucas A, Fu Q, Kent MP, Vega R, Matukumalli A, McWilliam S, Sclep G, Bryc K, Choi J, Gao H, Grefenstette JJ, Murdoch B, Stella A, Villa-Angulo R, Wright M, Aerts J, Jann O, Negrini R, Goddard ME, Hayes BJ, Bradley DG, Barbosa da Silva M, Lau LP, Liu GE, Lynn DJ, Panzitta F, Dodds KG. Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science. 2009;324:528–532. doi: 10.1126/science.1167936. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

