Quantitative phenotyping via deep barcode sequencing
- PMID: 19622793
- PMCID: PMC2765281
- DOI: 10.1101/gr.093955.109
Quantitative phenotyping via deep barcode sequencing
Abstract
Next-generation DNA sequencing technologies have revolutionized diverse genomics applications, including de novo genome sequencing, SNP detection, chromatin immunoprecipitation, and transcriptome analysis. Here we apply deep sequencing to genome-scale fitness profiling to evaluate yeast strain collections in parallel. This method, Barcode analysis by Sequencing, or "Bar-seq," outperforms the current benchmark barcode microarray assay in terms of both dynamic range and throughput. When applied to a complex chemogenomic assay, Bar-seq quantitatively identifies drug targets, with performance superior to the benchmark microarray assay. We also show that Bar-seq is well-suited for a multiplex format. We completely re-sequenced and re-annotated the yeast deletion collection using deep sequencing, found that approximately 20% of the barcodes and common priming sequences varied from expectation, and used this revised list of barcode sequences to improve data quality. Together, this new assay and analysis routine provide a deep-sequencing-based toolkit for identifying gene-environment interactions on a genome-wide scale.
Figures
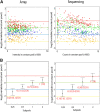


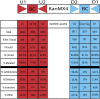
References
-
- Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5:433–438. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
