Molecular analysis of spring viraemia of carp virus in China: a fatal aquatic viral disease that might spread in East Asian
- PMID: 19623265
- PMCID: PMC2710009
- DOI: 10.1371/journal.pone.0006337
Molecular analysis of spring viraemia of carp virus in China: a fatal aquatic viral disease that might spread in East Asian
Abstract
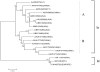
Spring viraemia of carp (SVC) is a fatal viral disease for cyprinid fish, which is caused by spring viraemia of carp virus (SVCV). To date, no SVC outbreak has been reported in China. Between 1998 and 2002, outbreaks of SVC were reported in ornamental and wild fish in Europe and America, imported from multiple sources including China. Based on phylogenetic analysis, the viral strain isolated from America was shown to be originated from Asia. These outbreaks not only resulted in huge economic losses, but also raise an interesting question as to whether SVCV really exists in China and if so, is it responsible for SVC outbreaks? From 2002 to 2006, we screened 6700 samples from ornamental fish farms using the cell culture method of the Office International des Epizooties (OIE), and further verified the presence of SVCV by ELISA and real-time quantitative RT-PCR. Two infected samples were found and the complete genome of SVCV was sequenced from one of the isolates, termed SVCV-C1. Several unique hallmarks of SVCV-C1 were identified, including six amino acid (KSLANA) insertion in the viral RNA-dependent RNA polymerase (L) protein and ten nucleotide insertion in the region between glycoprotein (G) and L genes in European SVCV strains. Phylogenetic tree analysis of the full-length G protein of selected SVCV isolates from the United Kingdom and United States revealed that G proteins could be classified into Ia and Id sub genogroups. The Ia sub genogroup can be further divided into newly defined sub genogroups Ia-A and Ia-B. The isolates derived from the United States and China including the SVCV-C1 belongs to in the Ia-A sub genogroup. The SVCV-C1 G protein shares more than 99% homology with the G proteins of the SVCV strains from England and the United States, making it difficult to compare their pathogenicity. Comparison of the predicted three-dimensional structure based on the published G protein sequences from five SVCV strains revealed that the main differences were in the loops of the pleckstrin homology domains. Since SVCV is highly pathogenic, we speculate that SVC may therefore pose a serious threat to farmed cyprinid fish in China.
Conflict of interest statement
Figures



References
-
- Walker PJ, BA, Calisher CH, et al. Family Rhabdoviridae. In: van Regenmortel MHV, Fauquet CM, Bishop DHL, Carstens EB, editors. The Seventh Report of the International Committee for Taxonomy of Viruses. CA: Academic Press San Diego; 2000. pp. 563–583. and 7 others.
-
- Ahne W, Bjorklund HV, Essbauer S, Fijan N, Kurath G, et al. Spring viremia of carp (SVC). Dis Aquat Organ. 2002;52:261–272. - PubMed
-
- Fijan N. Spring viremia of carp and other viral diseases of warm-water fish. In: Woo PTK, Bruno DW, editors. Fish diseases and disorders. Vol. 3. Oxon: CAB International; 1999. pp. 177–244.
-
- Bjorklund HV, Higman KH, Kurath G. The glycoprotein genes and gene junctions of the fish rhabdoviruses spring viremia of carp virus and hirame rhabdovirus: analysis of relationships with other rhabdoviruses. Virus Res. 1996;42:65–80. - PubMed
-
- Hoffmann B, Schutze H, Mettenleiter TC. Determination of the complete genomic sequence and analysis of the gene products of the virus of Spring Viremia of Carp, a fish rhabdovirus. Virus Res. 2002;84:89–100. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

