The evolutionary dynamics of the Helena retrotransposon revealed by sequenced Drosophila genomes
- PMID: 19624823
- PMCID: PMC3087515
- DOI: 10.1186/1471-2148-9-174
The evolutionary dynamics of the Helena retrotransposon revealed by sequenced Drosophila genomes
Abstract
Background: Several studies have shown that genomes contain a mixture of transposable elements, some of which are still active and others ancient relics that have degenerated. This is true for the non-LTR retrotransposon Helena, of which only degenerate sequences have been shown to be present in some species (Drosophila melanogaster), whereas putatively active sequences are present in others (D. simulans). Combining experimental and population analyses with the sequence analysis of the 12 Drosophila genomes, we have investigated the evolution of Helena, and propose a possible scenario for the evolution of this element.
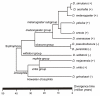
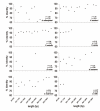
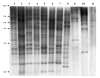
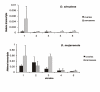
Results: We show that six species of Drosophila have the Helena transposable element at different stages of its evolution. The copy number is highly variable among these species, but most of them are truncated at the 5' ends and also harbor several internal deletions and insertions suggesting that they are inactive in all species, except in D. mojavensis in which quantitative RT-PCR experiments have identified a putative active copy.
Conclusion: Our data suggest that Helena was present in the common ancestor of the Drosophila genus, which has been vertically transmitted to the derived lineages, but that it has been lost in some of them. The wide variation in copy number and sequence degeneration in the different species suggest that the evolutionary dynamics of Helena depends on the genomic environment of the host species.
Figures


Similar articles
-
Evolutionary dynamics of the LTR retrotransposons roo and rooA inferred from twelve complete Drosophila genomes.BMC Evol Biol. 2009 Aug 18;9:205. doi: 10.1186/1471-2148-9-205. BMC Evol Biol. 2009. PMID: 19689787 Free PMC article.
-
Vertical inheritance and bursts of transposition have shaped the evolution of the BS non-LTR retrotransposon in Drosophila.Mol Genet Genomics. 2011 Jul;286(1):57-66. doi: 10.1007/s00438-011-0629-9. Epub 2011 May 27. Mol Genet Genomics. 2011. PMID: 21618036
-
High rate of DNA loss in the Drosophila melanogaster and Drosophila virilis species groups.Mol Biol Evol. 1998 Mar;15(3):293-302. doi: 10.1093/oxfordjournals.molbev.a025926. Mol Biol Evol. 1998. PMID: 9501496
-
Evolutionary forces generating sequence homogeneity and heterogeneity within retrotransposon families.Cytogenet Genome Res. 2005;110(1-4):383-91. doi: 10.1159/000084970. Cytogenet Genome Res. 2005. PMID: 16093690 Review.
-
Domesticated LTR-Retrotransposon gag-Related Gene (Gagr) as a Member of the Stress Response Network in Drosophila.Life (Basel). 2022 Mar 3;12(3):364. doi: 10.3390/life12030364. Life (Basel). 2022. PMID: 35330115 Free PMC article. Review.
Cited by
-
Losing identity: structural diversity of transposable elements belonging to different classes in the genome of Anopheles gambiae.BMC Genomics. 2012 Jun 22;13:272. doi: 10.1186/1471-2164-13-272. BMC Genomics. 2012. PMID: 22726298 Free PMC article.
-
VHICA, a New Method to Discriminate between Vertical and Horizontal Transposon Transfer: Application to the Mariner Family within Drosophila.Mol Biol Evol. 2016 Apr;33(4):1094-109. doi: 10.1093/molbev/msv341. Epub 2015 Dec 18. Mol Biol Evol. 2016. PMID: 26685176 Free PMC article.
-
Population genomics of transposable elements in Drosophila melanogaster.Mol Biol Evol. 2011 May;28(5):1633-44. doi: 10.1093/molbev/msq337. Epub 2010 Dec 16. Mol Biol Evol. 2011. PMID: 21172826 Free PMC article.
-
Drosophila embryogenesis scales uniformly across temperature in developmentally diverse species.PLoS Genet. 2014 Apr 24;10(4):e1004293. doi: 10.1371/journal.pgen.1004293. eCollection 2014 Apr. PLoS Genet. 2014. PMID: 24762628 Free PMC article.
-
Specific activation of an I-like element in Drosophila interspecific hybrids.Genome Biol Evol. 2014 Jun 24;6(7):1806-17. doi: 10.1093/gbe/evu141. Genome Biol Evol. 2014. PMID: 24966182 Free PMC article.
References
-
- Kaminker JS, Bergman CM, Kronmiller B, Carlson J, Svirskas R, Patel S, Frise E, Wheeler DA, Lewis SE, Rubin GM. et al.The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 2002;3(12):RESEARCH0084. doi: 10.1186/gb-2002-3-12-research0084. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

