Genome-wide analysis of the RpoN regulon in Geobacter sulfurreducens
- PMID: 19624843
- PMCID: PMC2725144
- DOI: 10.1186/1471-2164-10-331
Genome-wide analysis of the RpoN regulon in Geobacter sulfurreducens
Abstract
Background: The role of the RNA polymerase sigma factor RpoN in regulation of gene expression in Geobacter sulfurreducens was investigated to better understand transcriptional regulatory networks as part of an effort to develop regulatory modules for genome-scale in silico models, which can predict the physiological responses of Geobacter species during groundwater bioremediation or electricity production.
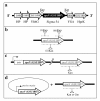
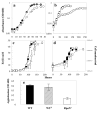
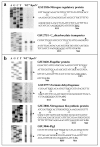
Results: An rpoN deletion mutant could not be obtained under all conditions tested. In order to investigate the regulon of the G. sulfurreducens RpoN, an RpoN over-expression strain was made in which an extra copy of the rpoN gene was under the control of a taclac promoter. Combining both the microarray transcriptome analysis and the computational prediction revealed that the G. sulfurreducens RpoN controls genes involved in a wide range of cellular functions. Most importantly, RpoN controls the expression of the dcuB gene encoding the fumarate/succinate exchanger, which is essential for cell growth with fumarate as the terminal electron acceptor in G. sulfurreducens. RpoN also controls genes, which encode enzymes for both pathways of ammonia assimilation that is predicted to be essential under all growth conditions in G. sulfurreducens. Other genes that were identified as part of the RpoN regulon using either the computational prediction or the microarray transcriptome analysis included genes involved in flagella biosynthesis, pili biosynthesis and genes involved in central metabolism enzymes and cytochromes involved in extracellular electron transfer to Fe(III), which are known to be important for growth in subsurface environment or electricity production in microbial fuel cells. The consensus sequence for the predicted RpoN-regulated promoter elements is TTGGCACGGTTTTTGCT.
Conclusion: The G. sulfurreducens RpoN is an essential sigma factor and a global regulator involved in a complex transcriptional network controlling a variety of cellular processes.
Figures


Similar articles
-
DNA microarray and proteomic analyses of the RpoS regulon in Geobacter sulfurreducens.J Bacteriol. 2006 Apr;188(8):2792-800. doi: 10.1128/JB.188.8.2792-2800.2006. J Bacteriol. 2006. PMID: 16585740 Free PMC article.
-
Genome-wide survey for PilR recognition sites of the metal-reducing prokaryote Geobacter sulfurreducens.Gene. 2010 Dec 1;469(1-2):31-44. doi: 10.1016/j.gene.2010.08.005. Epub 2010 Aug 12. Gene. 2010. PMID: 20708667
-
Computational prediction of RpoS and RpoD regulatory sites in Geobacter sulfurreducens using sequence and gene expression information.Gene. 2006 Dec 15;384:73-95. doi: 10.1016/j.gene.2006.06.025. Epub 2006 Jul 20. Gene. 2006. PMID: 17014972
-
Diversity of promoter elements in a Geobacter sulfurreducens mutant adapted to disruption in electron transfer.Funct Integr Genomics. 2009 Feb;9(1):15-25. doi: 10.1007/s10142-008-0094-7. Epub 2008 Aug 2. Funct Integr Genomics. 2009. PMID: 18677521 Review.
-
The Regulatory Functions of σ54 Factor in Phytopathogenic Bacteria.Int J Mol Sci. 2021 Nov 24;22(23):12692. doi: 10.3390/ijms222312692. Int J Mol Sci. 2021. PMID: 34884502 Free PMC article. Review.
Cited by
-
Genetic switches and related tools for controlling gene expression and electrical outputs of Geobacter sulfurreducens.J Ind Microbiol Biotechnol. 2016 Nov;43(11):1561-1575. doi: 10.1007/s10295-016-1836-5. Epub 2016 Sep 22. J Ind Microbiol Biotechnol. 2016. PMID: 27659960
-
Genome-wide gene regulation of biosynthesis and energy generation by a novel transcriptional repressor in Geobacter species.Nucleic Acids Res. 2010 Jan;38(3):810-21. doi: 10.1093/nar/gkp1085. Epub 2009 Nov 25. Nucleic Acids Res. 2010. PMID: 19939938 Free PMC article.
-
An advanced bioinformatics approach for analyzing RNA-seq data reveals sigma H-dependent regulation of competence genes in Listeria monocytogenes.BMC Genomics. 2016 Feb 16;17:115. doi: 10.1186/s12864-016-2432-9. BMC Genomics. 2016. PMID: 26880300 Free PMC article.
-
Influence of support materials on the electroactive behavior, structure and gene expression of wild type and GSU1771-deficient mutant of Geobacter sulfurreducens biofilms.Environ Sci Pollut Res Int. 2025 Jun;32(28):16740-16759. doi: 10.1007/s11356-024-33612-3. Epub 2024 May 17. Environ Sci Pollut Res Int. 2025. PMID: 38758442 Free PMC article.
-
Identification of metabolism pathways directly regulated by sigma(54) factor in Bacillus thuringiensis.Front Microbiol. 2015 May 12;6:407. doi: 10.3389/fmicb.2015.00407. eCollection 2015. Front Microbiol. 2015. PMID: 26029175 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

