Structural characterization of a viral NEIL1 ortholog unliganded and bound to abasic site-containing DNA
- PMID: 19625256
- PMCID: PMC2758016
- DOI: 10.1074/jbc.M109.021907
Structural characterization of a viral NEIL1 ortholog unliganded and bound to abasic site-containing DNA
Abstract
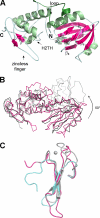
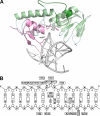
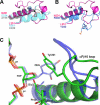
Endonuclease VIII (Nei) is a DNA glycosylase of the base excision repair pathway that recognizes and excises oxidized pyrimidines. We determined the crystal structures of a NEIL1 ortholog from the giant Mimivirus (MvNei1) unliganded and bound to DNA containing tetrahydrofuran (THF), which is the first structure of any Nei with an abasic site analog. The MvNei1 structures exhibit the same overall architecture as other enzymes of the Fpg/Nei family, which consists of two globular domains joined by a linker region. MvNei1 harbors a zincless finger, first described in human NEIL1, rather than the signature zinc finger generally found in the Fpg/Nei family. In contrast to Escherichia coli Nei, where a dramatic conformational change was observed upon binding DNA, the structure of MvNei1 bound to DNA does not reveal any substantial movement compared with the unliganded enzyme. A protein segment encompassing residues 217-245 in MvNei1 corresponds to the "missing loop" in E. coli Nei and the "alphaF-beta10 loop" in E. coli Fpg, which has been reported to be involved in lesion recognition. Interestingly, the corresponding loop in MvNei1 is ordered in both the unliganded and furan-bound structures, unlike other Fpg/Nei enzymes where the loop is generally ordered in the unliganded enzyme or in complexes with a lesion, and disordered otherwise. In the MvNei1.tetrahydrofuran complex a tyrosine located at the tip of the putative lesion recognition loop stacks against the furan ring; the tyrosine is predicted to adopt a different conformation to accommodate a modified base.
Figures


Similar articles
-
Human endonuclease VIII-like (NEIL) proteins in the giant DNA Mimivirus.DNA Repair (Amst). 2007 Nov;6(11):1629-41. doi: 10.1016/j.dnarep.2007.05.011. Epub 2007 Jul 12. DNA Repair (Amst). 2007. PMID: 17627905 Free PMC article.
-
Structural and biochemical studies of a plant formamidopyrimidine-DNA glycosylase reveal why eukaryotic Fpg glycosylases do not excise 8-oxoguanine.DNA Repair (Amst). 2012 Sep 1;11(9):714-25. doi: 10.1016/j.dnarep.2012.06.004. Epub 2012 Jul 11. DNA Repair (Amst). 2012. PMID: 22789755 Free PMC article.
-
Structure of the uncomplexed DNA repair enzyme endonuclease VIII indicates significant interdomain flexibility.Nucleic Acids Res. 2005 Sep 6;33(15):5006-16. doi: 10.1093/nar/gki796. Print 2005. Nucleic Acids Res. 2005. PMID: 16145054 Free PMC article.
-
The Fpg/Nei family of DNA glycosylases: substrates, structures, and search for damage.Prog Mol Biol Transl Sci. 2012;110:71-91. doi: 10.1016/B978-0-12-387665-2.00004-3. Prog Mol Biol Transl Sci. 2012. PMID: 22749143 Free PMC article. Review.
-
Structural characterization of the Fpg family of DNA glycosylases.DNA Repair (Amst). 2003 Aug 12;2(8):839-62. doi: 10.1016/s1568-7864(03)00084-3. DNA Repair (Amst). 2003. PMID: 12893082 Review.
Cited by
-
Using shifts in amino acid frequency and substitution rate to identify latent structural characters in base-excision repair enzymes.PLoS One. 2011;6(10):e25246. doi: 10.1371/journal.pone.0025246. Epub 2011 Oct 6. PLoS One. 2011. PMID: 21998646 Free PMC article.
-
Non-specific DNA binding interferes with the efficient excision of oxidative lesions from chromatin by the human DNA glycosylase, NEIL1.DNA Repair (Amst). 2010 Feb 4;9(2):134-43. doi: 10.1016/j.dnarep.2009.11.005. Epub 2009 Dec 11. DNA Repair (Amst). 2010. PMID: 20005182 Free PMC article.
-
A novel link to base excision repair?Trends Biochem Sci. 2010 May;35(5):247-52. doi: 10.1016/j.tibs.2010.01.003. Epub 2010 Feb 19. Trends Biochem Sci. 2010. PMID: 20172733 Free PMC article.
-
Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair.Proc Natl Acad Sci U S A. 2016 Jul 12;113(28):7792-7. doi: 10.1073/pnas.1604591113. Epub 2016 Jun 27. Proc Natl Acad Sci U S A. 2016. PMID: 27354518 Free PMC article.
-
A Low-Activity Polymorphic Variant of Human NEIL2 DNA Glycosylase.Int J Mol Sci. 2022 Feb 17;23(4):2212. doi: 10.3390/ijms23042212. Int J Mol Sci. 2022. PMID: 35216329 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

