MD simulations of ligand-bound and ligand-free aptamer: molecular level insights into the binding and switching mechanism of the add A-riboswitch
- PMID: 19625387
- PMCID: PMC2743061
- DOI: 10.1261/rna.1675809
MD simulations of ligand-bound and ligand-free aptamer: molecular level insights into the binding and switching mechanism of the add A-riboswitch
Erratum in
- RNA. 2009 Nov;15(11):2072
Abstract
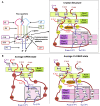
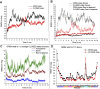
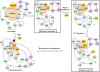
Riboswitches are structural cis-acting genetic regulatory elements in 5' UTRs of mRNAs, consisting of an aptamer domain that regulates the behavior of an expression platform in response to its recognition of, and binding to, specific ligands. While our understanding of the ligand-bound structure of the aptamer domain of the adenine riboswitches is based on crystal structure data and is well characterized, understanding of the structure and dynamics of the ligand-free aptamer is limited to indirect inferences from physicochemical probing experiments. Here we report the results of 15-nsec-long explicit-solvent molecular dynamics simulations of the add A-riboswitch crystal structure (1Y26), both in the adenine-bound (CLOSED) state and in the adenine-free (OPEN) state. Root-mean-square deviation, root-mean-square fluctuation, dynamic cross-correlation, and backbone torsion angle analyses are carried out on the two trajectories. These, along with solvent accessible surface area analysis of the two average structures, are benchmarked against available experimental data and are shown to constitute the basis for obtaining reliable insights into the molecular level details of the binding and switching mechanism. Our analysis reveals the interaction network responsible for, and conformational changes associated with, the communication between the binding pocket and the expression platform. It further highlights the significance of a, hitherto unreported, noncanonical W:H trans base pairing between A73 and A24, in the OPEN state, and also helps us to propose a possibly crucial role of U51 in the context of ligand binding and ligand discrimination.
Figures




Similar articles
-
Molecular dynamics simulation of the binding process of ligands to the add adenine riboswitch aptamer.Phys Rev E. 2019 Aug;100(2-1):022412. doi: 10.1103/PhysRevE.100.022412. Phys Rev E. 2019. PMID: 31574664
-
Exploring the Binding Process of Cognate Ligand to Add Adenine Riboswitch Aptamer by Using Explicit Solvent Molecular Dynamics (MD) Simulation.Methods Mol Biol. 2023;2568:103-122. doi: 10.1007/978-1-0716-2687-0_7. Methods Mol Biol. 2023. PMID: 36227564
-
Role of ligand binding in structural organization of add A-riboswitch aptamer: a molecular dynamics simulation.J Biomol Struct Dyn. 2011 Oct;29(2):403-16. doi: 10.1080/07391102.2011.10507394. J Biomol Struct Dyn. 2011. PMID: 21875158
-
Linking aptamer-ligand binding and expression platform folding in riboswitches: prospects for mechanistic modeling and design.Wiley Interdiscip Rev RNA. 2015 Nov-Dec;6(6):631-50. doi: 10.1002/wrna.1300. Epub 2015 Sep 11. Wiley Interdiscip Rev RNA. 2015. PMID: 26361734 Free PMC article. Review.
-
Riboswitch Mechanisms for Regulation of P1 Helix Stability.Int J Mol Sci. 2024 Oct 4;25(19):10682. doi: 10.3390/ijms251910682. Int J Mol Sci. 2024. PMID: 39409011 Free PMC article. Review.
Cited by
-
Potential effects of metal ion induced two-state allostery on the regulatory mechanism of add adenine riboswitch.Commun Biol. 2022 Oct 22;5(1):1120. doi: 10.1038/s42003-022-04096-z. Commun Biol. 2022. PMID: 36273041 Free PMC article.
-
The purine riboswitch as a model system for exploring RNA biology and chemistry.Biochim Biophys Acta. 2014 Oct;1839(10):919-930. doi: 10.1016/j.bbagrm.2014.02.014. Epub 2014 Feb 28. Biochim Biophys Acta. 2014. PMID: 24590258 Free PMC article. Review.
-
DFT and MD study of the divalent-cation-mediated interaction of ochratoxin A with DNA nucleosides.J Mol Model. 2014 Jun;20(6):2274. doi: 10.1007/s00894-014-2274-9. Epub 2014 May 27. J Mol Model. 2014. PMID: 24863531
-
Role of the adenine ligand on the stabilization of the secondary and tertiary interactions in the adenine riboswitch.J Mol Biol. 2010 Mar 12;396(5):1422-38. doi: 10.1016/j.jmb.2009.12.024. Epub 2009 Dec 21. J Mol Biol. 2010. PMID: 20026131 Free PMC article.
-
Single-molecule chemical denaturation of riboswitches.Nucleic Acids Res. 2013 Apr;41(7):4253-65. doi: 10.1093/nar/gkt128. Epub 2013 Feb 27. Nucleic Acids Res. 2013. PMID: 23446276 Free PMC article.
References
-
- Bansal M, Bhattacharyya D, Ravi B. NUPARM and NUCGEN: Software for analysis and generation of sequence dependent nucleic acid structures. Comput Appl Biosci. 1995;11:281–287. - PubMed
-
- Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. - PubMed
-
- Bhattacharyya D, Koripella SC, Mitra A, Rajendran VB, Sinha B. Theoretical analysis of noncanonical base pairing interactions in RNA molecules. J Biosci. 2007;32:809–825. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
