Human immunodeficiency virus type 1 V1-to-V5 envelope variants from the chronic phase of infection use CCR5 and fuse more efficiently than those from early after infection
- PMID: 19625411
- PMCID: PMC2748008
- DOI: 10.1128/JVI.00925-09
Human immunodeficiency virus type 1 V1-to-V5 envelope variants from the chronic phase of infection use CCR5 and fuse more efficiently than those from early after infection
Abstract
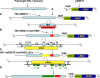
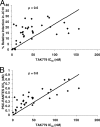
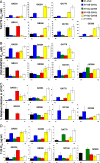
Human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein modifications over the course of infection have been associated with coreceptor switching and antibody neutralization resistance, but the effect of the changes on replication and host cell receptor usage remains unclear. To examine this question, unique early- and chronic-stage infection envelope V1-to V5 (V1-V5) segments from eight HIV-1 subtype A-infected subjects were incorporated into an isogenic background to construct replication-competent recombinant viruses. In all subjects, viruses with chronic-infection V1-V5 segments showed greater replication capacity than those with early-infection V1-V5 domains in cell lines with high levels of both the CD4 and the CCR5 receptors. Viruses with chronic-infection V1-V5s demonstrated a significantly increased ability to replicate in cells with low CCR5 receptor levels and greater resistance to CCR5 receptor and fusion inhibitors compared to those with early-infection V1-V5 segments. These properties were associated with sequence changes in the envelope V1-V3 segments. Viruses with the envelope segments from the two infection time points showed no significant difference in their ability to infect cells with low CD4 receptor densities, in their sensitivity to soluble CD4, or in their replication capacity in monocyte-derived macrophages. Our results suggest that envelope changes, primarily in the V1-V3 domains, increase both the ability to use the CCR5 receptor and fusion kinetics. Thus, envelope modifications over time within a host potentially enhance replication capacity.
Figures
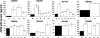


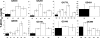
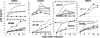
References
-
- Alkhatib, G., M. Locati, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1997. HIV-1 coreceptor activity of CCR5 and its inhibition by chemokines: independence from G protein signaling and importance of coreceptor downmodulation. Virology 234:340-348. - PubMed
-
- Baba, M., O. Nishimura, N. Kanzaki, M. Okamoto, H. Sawada, Y. Iizawa, M. Shiraishi, Y. Aramaki, K. Okonogi, Y. Ogawa, K. Meguro, and M. Fujino. 1999. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc. Natl. Acad. Sci. USA 96:5698-5703. - PMC - PubMed
-
- Ball, S. C., A. Abraha, K. R. Collins, A. J. Marozsan, H. Baird, M. E. Quinones-Mateu, A. Penn-Nicholson, M. Murray, N. Richard, M. Lobritz, P. A. Zimmerman, T. Kawamura, A. Blauvelt, and E. J. Arts. 2003. Comparing the ex vivo fitness of CCR5-tropic human immunodeficiency virus type 1 isolates of subtypes B and C. J. Virol. 77:1021-1038. - PMC - PubMed
-
- Blaak, H., M. Brouwer, L. J. Ran, F. de Wolf, and H. Schuitemaker. 1998. In vitro replication kinetics of human immunodeficiency virus type 1 (HIV-1) variants in relation to virus load in long-term survivors of HIV-1 infection. J. Infect. Dis. 177:600-610. - PubMed
-
- Brenchley, J. M., T. W. Schacker, L. E. Ruff, D. A. Price, J. H. Taylor, G. J. Beilman, P. L. Nguyen, A. Khoruts, M. Larson, A. T. Haase, and D. C. Douek. 2004. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J. Exp. Med. 200:749-759. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

