Evaluation of the Abbott investigational use only RealTime hepatitis C virus (HCV) assay and comparison to the Roche TaqMan HCV analyte-specific reagent assay
- PMID: 19625475
- PMCID: PMC2738061
- DOI: 10.1128/JCM.02329-08
Evaluation of the Abbott investigational use only RealTime hepatitis C virus (HCV) assay and comparison to the Roche TaqMan HCV analyte-specific reagent assay
Abstract
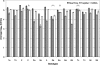
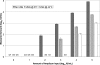
The accurate and sensitive measurement of hepatitis C virus (HCV) RNA is essential for the clinical management and treatment of infected patients and as a research tool for studying the biology of HCV infection. We evaluated the linearity, reproducibility, precision, limit of detection, and concordance of viral genotype quantitation of the Abbott investigational use only RealTime HCV (RealTime) assay using the Abbott m2000 platform and compared the results to those of the Roche TaqMan Analyte-Specific Reagent (TaqMan) and Bayer Versant HCV bDNA 3.0 assay. Comparison of 216 samples analyzed by RealTime and TaqMan assays produced the following Deming regression equation: RealTime = 0.940 (TaqMan) + 0.175 log(10) HCV RNA IU/ml. The average difference between the assays was 0.143 log(10) RNA IU/ml and was consistent across RealTime's dynamic range of nearly 7 log(10) HCV RNA IU/ml. There was no significant difference between genotypes among these samples. The limit of detection using eight replicates of the World Health Organization HCV standard was determined to be 7.74 HCV RNA IU/ml by probit analysis. Replicate measurements of commercial genotype panels were significantly higher than TaqMan measurements for most samples and showed that the RealTime assay is able to detect all genotypes with no bias. Additionally, we showed that the amplicon generated by the widely used Roche COBAS Amplicor Hepatitis C Virus Test, version 2.0, can act as a template in the RealTime assay, but potential cross-contamination could be mitigated by treatment with uracil-N-glycosylase. In conclusion, the RealTime assay accurately measured HCV viral loads over a broad dynamic range, with no significant genotype bias.
Figures

References
-
- Bland, J. M., and D. G. Altman. 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1307-310. - PubMed
-
- Braun, P., R. Ehret, F. Wiesmann, F. Zabbai, M. Knickmann, R Kühn, S. Thamm, G. Warnat, and H. Knechten. 2007. Comparison of four commercial quantitative HIV-1 assays for viral load monitoring in clinical daily routine. Clin. Chem. Lab. Med. 4593-99. - PubMed
-
- Elbeik, T., R. Dalessandro, R. A. Loftus, and S. Beringer. 2007. HIV-1 and HCV viral load cost models for bDNA: 440 Molecular System versus real-time PCR AmpliPrep/TaqMan test. Expert Rev. Mol. Diagn. 7723-753. - PubMed
-
- Halfon, P., G. Pénaranda, M. Bourlière, H. Khiri, M.-F. Masseyeff, and D. Ouzan. 2006. Assessment of early virological response to antiviral therapy by comparing four assays for HCV RNA quantitation using the international unit standard: implications for clinical management of patients with chronic hepatitis C virus infection. J. Med. Virol. 78208-215. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

