Retina-specific GTPase accelerator RGS11/G beta 5S/R9AP is a constitutive heterotrimer selectively targeted to mGluR6 in ON-bipolar neurons
- PMID: 19625520
- PMCID: PMC2731308
- DOI: 10.1523/JNEUROSCI.1367-09.2009
Retina-specific GTPase accelerator RGS11/G beta 5S/R9AP is a constitutive heterotrimer selectively targeted to mGluR6 in ON-bipolar neurons
Abstract
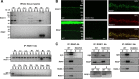
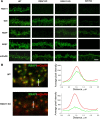
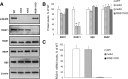
Members of the R7 family of the regulators of G-protein signaling (R7 RGS) proteins form multi-subunit complexes that play crucial roles in processing the light responses of retinal neurons. The disruption of these complexes has been shown to lead to the loss of temporal resolution in retinal photoreceptors and deficient synaptic transmission to downstream neurons. Despite the well established role of one member of this family, RGS9-1, in controlling vertebrate phototransduction, the roles and organizational principles of other members in the retina are poorly understood. Here we investigate the composition, localization, and function of complexes containing RGS11, the closest homolog of RGS9-1. We find that RGS11 forms a novel obligatory trimeric complex with the short splice isoform of the type 5 G-protein beta subunit (G beta 5) and the RGS9 anchor protein (R9AP). The complex is expressed exclusively in the dendritic tips of ON-bipolar cells in which its localization is accomplished through a direct association with mGluR6, the glutamate receptor essential for the ON-bipolar light response. Although association with both R9AP and mGluR6 contributed to the proteolytic stabilization of the complex, postsynaptic targeting of RGS11 was not determined by its membrane anchor R9AP. Electrophysiological recordings of the light response in mouse rod ON-bipolar cells reveal that the genetic elimination of RGS11 has little effect on the deactivation of G alpha(o) in dark-adapted cells or during adaptation to background light. These results suggest that the deactivation of mGluR6 cascade during the light response may require the contribution of multiple GTPase activating proteins.
Figures






Similar articles
-
Membrane anchor R9AP potentiates GTPase-accelerating protein activity of RGS11 x Gbeta5 complex and accelerates inactivation of the mGluR6-G(o) signaling.J Biol Chem. 2010 Feb 12;285(7):4781-7. doi: 10.1074/jbc.M109.058511. Epub 2009 Dec 11. J Biol Chem. 2010. PMID: 20007977 Free PMC article.
-
R9AP stabilizes RGS11-G beta5 and accelerates the early light response of ON-bipolar cells.Vis Neurosci. 2010 Mar;27(1-2):9-17. doi: 10.1017/S0952523809990319. Epub 2010 Jan 26. Vis Neurosci. 2010. PMID: 20100392 Free PMC article.
-
Gbeta5-RGS complexes co-localize with mGluR6 in retinal ON-bipolar cells.Eur J Neurosci. 2007 Nov;26(10):2899-905. doi: 10.1111/j.1460-9568.2007.05867.x. Eur J Neurosci. 2007. PMID: 18001285 Free PMC article.
-
RGS Protein Regulation of Phototransduction.Prog Mol Biol Transl Sci. 2015;133:31-45. doi: 10.1016/bs.pmbts.2015.02.004. Epub 2015 Apr 16. Prog Mol Biol Transl Sci. 2015. PMID: 26123301 Free PMC article. Review.
-
The R7 RGS protein family: multi-subunit regulators of neuronal G protein signaling.Cell Biochem Biophys. 2009;54(1-3):33-46. doi: 10.1007/s12013-009-9052-9. Epub 2009 Jun 12. Cell Biochem Biophys. 2009. PMID: 19521673 Free PMC article. Review.
Cited by
-
Transience of the Retinal Output Is Determined by a Great Variety of Circuit Elements.Cells. 2022 Feb 25;11(5):810. doi: 10.3390/cells11050810. Cells. 2022. PMID: 35269432 Free PMC article. Review.
-
RGS9 knockout causes a short delay in light responses of ON-bipolar cells.PLoS One. 2011;6(11):e27573. doi: 10.1371/journal.pone.0027573. Epub 2011 Nov 11. PLoS One. 2011. PMID: 22096596 Free PMC article.
-
Murine Retina Outer Plexiform Layer Development and Transcriptome Analysis of Pre-Synapses in Photoreceptors.Life (Basel). 2024 Sep 2;14(9):1103. doi: 10.3390/life14091103. Life (Basel). 2024. PMID: 39337887 Free PMC article.
-
Molecular mechanisms underlying selective synapse formation of vertebrate retinal photoreceptor cells.Cell Mol Life Sci. 2020 Apr;77(7):1251-1266. doi: 10.1007/s00018-019-03324-w. Epub 2019 Oct 4. Cell Mol Life Sci. 2020. PMID: 31586239 Free PMC article. Review.
-
Illuminating the multifaceted roles of neurotransmission in shaping neuronal circuitry.Neuron. 2014 Sep 17;83(6):1303-1318. doi: 10.1016/j.neuron.2014.08.029. Neuron. 2014. PMID: 25233313 Free PMC article. Review.
References
-
- Anderson GR, Semenov A, Song JH, Martemyanov KA. The membrane anchor R7BP controls the proteolytic stability of the striatal specific RGS protein, RGS9–2. J Biol Chem. 2007a;282:4772–4781. - PubMed
-
- Anderson GR, Lujan R, Semenov A, Pravetoni M, Posokhova EN, Song JH, Uversky V, Chen CK, Wickman K, Martemyanov KA. Expression and localization of RGS9–2/G 5/R7BP complex in vivo is set by dynamic control of its constitutive degradation by cellular cysteine proteases. J Neurosci. 2007b;27:14117–14127. - PMC - PubMed
-
- Ballon DR, Flanary PL, Gladue DP, Konopka JB, Dohlman HG, Thorner J. DEP-domain-mediated regulation of GPCR signaling responses. Cell. 2006;126:1079–1093. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
