Characterization and distribution of retrotransposons and simple sequence repeats in the bovine genome
- PMID: 19625614
- PMCID: PMC2722308
- DOI: 10.1073/pnas.0901282106
Characterization and distribution of retrotransposons and simple sequence repeats in the bovine genome
Abstract
Interspersed repeat composition and distribution in mammals have been best characterized in the human and mouse genomes. The bovine genome contains typical eutherian mammal repeats, but also has a significant number of long interspersed nuclear element RTE (BovB) elements proposed to have been horizontally transferred from squamata. Our analysis of the BovB repeats has indicated that only a few of them are currently likely to retrotranspose in cattle. However, bovine L1 repeats (L1 BT) have many likely active copies. Comparison of substitution rates for BovB and L1 BT indicates that L1 BT is a younger repeat family than BovB. In contrast to mouse and human, L1 occurrence is not negatively correlated with G+C content. However, BovB, Bov A2, ART2A, and Bov-tA are negatively correlated with G+C, although Bov-tAs correlation is weaker. Also, by performing genome wide correlation analysis of interspersed and simple sequence repeats, we have identified genome territories by repeat content that appear to define ancestral vs. ruminant-specific genomic regions. These ancestral regions, enriched with L2 and MIR repeats, are largely conserved between bovine and human.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

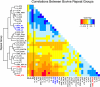

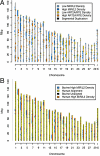
References
-
- Smit AF. The origin of interspersed repeats in the human genome. Curr Opin Genet Dev. 1996;6:743–748. - PubMed
-
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Jurka J, Kapitonov VV, Kohany O, Jurka MV. Repetitive sequences in complex genomes: Structure and evolution. Annu Rev Genom Hum G. 2007;8:241–259. - PubMed
-
- Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

