The preferentially expressed antigen in melanoma (PRAME) inhibits myeloid differentiation in normal hematopoietic and leukemic progenitor cells
- PMID: 19625708
- PMCID: PMC2759652
- DOI: 10.1182/blood-2008-07-170282
The preferentially expressed antigen in melanoma (PRAME) inhibits myeloid differentiation in normal hematopoietic and leukemic progenitor cells
Abstract
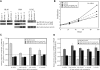
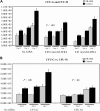
The preferentially expressed antigen in melanoma (PRAME) is expressed in several hematologic malignancies, but either is not expressed or is expressed at only low levels in normal hematopoietic cells, making it a target for cancer therapy. PRAME is a tumor-associated antigen and has been described as a corepressor of retinoic acid signaling in solid tumor cells, but its function in hematopoietic cells is unknown. PRAME mRNA expression increased with chronic myeloid leukemia (CML) disease progression and its detection in late chronic-phase CML patients before tyrosine kinase inhibitor therapy was associated with poorer therapeutic responses and ABL tyrosine kinase domain point mutations. In leukemia cell lines, PRAME protein expression inhibited granulocytic differentiation only in cell lines that differentiate along this lineage after all-trans retinoic acid (ATRA) exposure. Forced PRAME expression in normal hematopoietic progenitors, however, inhibited myeloid differentiation both in the presence and absence of ATRA, and this phenotype was reversed when PRAME was silenced in primary CML progenitors. These observations suggest that PRAME inhibits myeloid differentiation in certain myeloid leukemias, and that its function in these cells is lineage and phenotype dependent. Lastly, these observations suggest that PRAME is a target for both prognostic and therapeutic applications.
Figures



References
-
- Ikeda H, Lethe B, Lehmann F, et al. Characterization of an antigen that is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity. 1997;6(2):199–208. - PubMed
-
- van Baren N, Chambost H, Ferrant A, et al. PRAME, a gene encoding an antigen recognized on a human melanoma by cytolytic T cells, is expressed in acute leukaemia cells. Br J Haematol. 1998;102(5):1376–1379. - PubMed
-
- Steinbach D, Hermann J, Viehmann S, Zintl F, Gruhn B. Clinical implications of PRAME gene expression in childhood acute myeloid leukemia. Cancer Genet Cytogenet. 2002;133(2):118–123. - PubMed
-
- Epping MT, Wang L, Edel MJ, Carlee L, Hernandez M, Bernards R. The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell. 2005;122(6):835–847. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

