Capturing single molecules of immunoglobulin and ricin with an aptamer-encoded glass nanopore
- PMID: 19627120
- PMCID: PMC3009471
- DOI: 10.1021/ac9006705
Capturing single molecules of immunoglobulin and ricin with an aptamer-encoded glass nanopore
Abstract
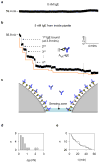
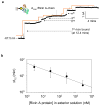
Nanopore-based single-molecule biosensors have been extensively studied. Protein pores that have receptors attached to them are target-selective, but their real-time applications are limited by the fragility of the lipid membrane into which the protein pores are embedded. Synthetic nanopores are more stable and provide flexible pore sizes, but the selectivity is low when detecting in the translocation mode. In spite of modifications with probing molecules, such as antibodies, to potentiate specific targeting, these nanopores fail to bind individual target molecules. Distinguishing between binding and translocation blocks remains unsolved. Here, we propose an aptamer-encoded nanopore that overcomes these challenges. Aptamers are well-known probing oligonucleotides that have high sensitivity and selectivity. In contrast to antibodies, aptamers are much smaller than their targets, rendering target blockades in the nanopore much more distinguishable. We used aptamer-encoded nanopores to detect single molecules of immunoglobulin E and the bioterrorist agent ricin, sequentially captured by the immobilized aptamer in the sensing zone of the pore. The functional nanopore also probed sequence-dependent aptamer-protein interactions. These findings will facilitate the development of a universal nanopore for multitarget detection.
Conflict of interest statement
Figures



Similar articles
-
Aptamer-encoded nanopore for ultrasensitive detection of bioterrorist agent ricin at single-molecule resolution.Annu Int Conf IEEE Eng Med Biol Soc. 2009;2009:6699-702. doi: 10.1109/IEMBS.2009.5333281. Annu Int Conf IEEE Eng Med Biol Soc. 2009. PMID: 19964179 Free PMC article. Review.
-
Single-Molecule Investigation of the Protein-Aptamer Interactions and Sensing Application Inside the Single Glass Nanopore.Anal Chem. 2022 Dec 20;94(50):17405-17412. doi: 10.1021/acs.analchem.2c02660. Epub 2022 Dec 7. Anal Chem. 2022. PMID: 36475604
-
Ultrasensitive and regenerable nanopore sensing based on target induced aptamer dissociation.Biosens Bioelectron. 2020 Mar 15;152:112011. doi: 10.1016/j.bios.2020.112011. Epub 2020 Jan 8. Biosens Bioelectron. 2020. PMID: 32056734
-
Single molecule sensing by nanopores and nanopore devices.Analyst. 2010 Mar;135(3):441-51. doi: 10.1039/b907735a. Epub 2009 Dec 22. Analyst. 2010. PMID: 20174694 Free PMC article. Review.
-
Recent Advances in Aptamer-Based Nanopore Sensing at Single-Molecule Resolution.Chem Asian J. 2022 Aug 15;17(16):e202200364. doi: 10.1002/asia.202200364. Epub 2022 Jun 23. Chem Asian J. 2022. PMID: 35644914 Review.
Cited by
-
Selective Detection of Protein Homologues in Serum Using an OmpG Nanopore.Anal Chem. 2015 Nov 3;87(21):11143-9. doi: 10.1021/acs.analchem.5b03350. Epub 2015 Oct 23. Anal Chem. 2015. PMID: 26451707 Free PMC article.
-
Resolved single-molecule detection of individual species within a mixture of anti-biotin antibodies using an engineered monomeric nanopore.ACS Nano. 2015 Feb 24;9(2):1089-98. doi: 10.1021/nn506606e. Epub 2015 Jan 22. ACS Nano. 2015. PMID: 25575121 Free PMC article.
-
Ultrasensitive mycotoxin detection by STING sensors.Biosens Bioelectron. 2010 Oct 15;26(2):333-7. doi: 10.1016/j.bios.2010.08.016. Epub 2010 Aug 19. Biosens Bioelectron. 2010. PMID: 20829024 Free PMC article.
-
Electrically facilitated translocation of protein through solid nanopore.Nanoscale Res Lett. 2014 Mar 24;9(1):140. doi: 10.1186/1556-276X-9-140. Nanoscale Res Lett. 2014. PMID: 24661490 Free PMC article.
-
When less is more in a nanopore.Nat Nanotechnol. 2012 Apr 5;7(4):212-3. doi: 10.1038/nnano.2012.48. Nat Nanotechnol. 2012. PMID: 22481488 No abstract available.
References
-
- Bayley H, Cremer PS. Nature. 2001;413:226–30. - PubMed
-
- Bezrukov SM, Vodyanoy I, Parsegian VA. Nature. 1994;370:279–81. - PubMed
-
- Gu LQ, Braha O, Conlan S, Cheley S, Bayley H. Nature. 1999;398:686–90. - PubMed
-
- Movileanu L, Howorka S, Braha O, Bayley H. Nature Biotechnology. 2000;18:1091–95. - PubMed
-
- Siwy Z, Trofin L, Kohli P, Baker LA, Trautmann C, Martin CR. Journal of the American Chemical Society. 2005;127:5000–01. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

