Genetic diversity of the E protein of dengue type 3 virus
- PMID: 19627608
- PMCID: PMC2720943
- DOI: 10.1186/1743-422X-6-113
Genetic diversity of the E protein of dengue type 3 virus
Abstract
Background: Dengue is the most important arbovirus disease in tropical and subtropical countries. The viral envelope (E) protein is responsible for cell receptor binding and is the main target of neutralizing antibodies. The aim of this study was to analyze the diversity of the E protein gene of DENV-3. E protein gene sequences of 20 new viruses isolated in Ribeirao Preto, Brazil, and 427 sequences retrieved from GenBank were aligned for diversity and phylogenetic analysis.
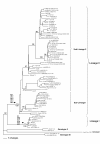
Results: Comparison of the E protein gene sequences revealed the presence of 47 variable sites distributed in the protein; most of those amino acids changes are located on the viral surface. The phylogenetic analysis showed the distribution of DENV-3 in four genotypes. Genotypes I, II and III revealed internal groups that we have called lineages and sub-lineages. All amino acids that characterize a group (genotype, lineage, or sub-lineage) are located in the 47 variable sites of the E protein.
Conclusion: Our results provide information about the most frequent amino acid changes and diversity of the E protein of DENV-3.
Figures



Similar articles
-
Phylogenetic relationship of dengue virus type 3 isolated in Brazil and Paraguay and global evolutionary divergence dynamics.Virol J. 2012 Jun 20;9:124. doi: 10.1186/1743-422X-9-124. Virol J. 2012. PMID: 22716071 Free PMC article.
-
Molecular surveillance of dengue in Minas Gerais provides insights on dengue virus 1 and 4 circulation in Brazil.J Med Virol. 2017 Jun;89(6):966-973. doi: 10.1002/jmv.24729. Epub 2016 Dec 7. J Med Virol. 2017. PMID: 27926790
-
Phylogenetic history demonstrates two different lineages of dengue type 1 virus in Colombia.Virol J. 2010 Sep 14;7:226. doi: 10.1186/1743-422X-7-226. Virol J. 2010. PMID: 20836894 Free PMC article.
-
Dengue virus type 4 phylogenetics in Brazil 2011: looking beyond the veil.PLoS Negl Trop Dis. 2011 Dec;5(12):e1439. doi: 10.1371/journal.pntd.0001439. Epub 2011 Dec 27. PLoS Negl Trop Dis. 2011. PMID: 22216365 Free PMC article.
-
Insights of the genetic diversity of DENV-1 detected in Brazil in 25 years: Analysis of the envelope domain III allows lineages characterization.Infect Genet Evol. 2015 Aug;34:126-36. doi: 10.1016/j.meegid.2015.07.007. Epub 2015 Jul 6. Infect Genet Evol. 2015. PMID: 26160541
Cited by
-
Dengue virus 2 American-Asian genotype identified during the 2006/2007 outbreak in Piauí, Brazil reveals a Caribbean route of introduction and dissemination of dengue virus in Brazil.PLoS One. 2014 Aug 15;9(8):e104516. doi: 10.1371/journal.pone.0104516. eCollection 2014. PLoS One. 2014. PMID: 25127366 Free PMC article.
-
Characterization of a Novel Dengue Serotype 4 Virus-Specific Neutralizing Epitope on the Envelope Protein Domain III.PLoS One. 2015 Oct 2;10(10):e0139741. doi: 10.1371/journal.pone.0139741. eCollection 2015. PLoS One. 2015. PMID: 26430770 Free PMC article.
-
Development and characterization of a reverse genetic system for studying dengue virus serotype 3 strain variation and neutralization.PLoS Negl Trop Dis. 2012;6(2):e1486. doi: 10.1371/journal.pntd.0001486. Epub 2012 Feb 28. PLoS Negl Trop Dis. 2012. PMID: 22389731 Free PMC article.
-
Challenges in dengue research: A computational perspective.Evol Appl. 2017 Nov 5;11(4):516-533. doi: 10.1111/eva.12554. eCollection 2018 Apr. Evol Appl. 2017. PMID: 29636803 Free PMC article. Review.
-
Mosquitoes infected with dengue viruses in Brazil.Virol J. 2010 Jul 12;7:152. doi: 10.1186/1743-422X-7-152. Virol J. 2010. PMID: 20624314 Free PMC article.
References
-
- WHO World Health Organization. Dengue and Dengue Haemorrhagic Fever. Fact Sheet No. 117. Geneva. 2002.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical

