RIG-I is cleaved during picornavirus infection
- PMID: 19628239
- PMCID: PMC2743091
- DOI: 10.1016/j.virol.2009.06.045
RIG-I is cleaved during picornavirus infection
Abstract
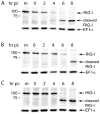
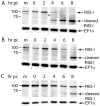
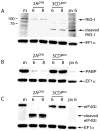
The innate immune system senses RNA virus infections through membrane-bound Toll-like receptors or the cytoplasmic proteins RIG-I and MDA-5. RIG-I is believed to recognize the 5'-triphosphate present on many viral RNAs, and hence is important for sensing infections by paramyxoviruses, influenza viruses, rhabdoviruses, and flaviviruses. MDA-5 recognizes dsRNA, and senses infection with picornaviruses, whose RNA 5'-ends are linked to a viral protein, VPg, not a 5'-triphosphate. We previously showed that MDA-5 is degraded in cells infected with different picornaviruses, and suggested that such cleavage might be a mechanism to antagonize production of type I IFN in response to viral infection. Here we examined the state of RIG-I during picornavirus infection. RIG-I is degraded in cells infected with poliovirus, rhinoviruses, echovirus, and encephalomyocarditis virus. In contrast to MDA-5, cleavage of RIG-I is not accomplished by cellular caspases or the proteasome. Rather, the viral proteinase 3C(pro) cleaves RIG-I, both in vitro and in cells. Cleavage of RIG-I during picornavirus infection may constitute another mechanism for attenuating the innate response to viral infection.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

