MicroRNAs with a nucleolar location
- PMID: 19628621
- PMCID: PMC2743059
- DOI: 10.1261/rna.1470409
MicroRNAs with a nucleolar location
Abstract
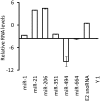
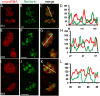
There is increasing evidence that noncoding RNAs play a functional role in the nucleus. We previously reported that the microRNA (miRNA), miR-206, is concentrated in the nucleolus of rat myoblasts, as well as in the cytoplasm as expected. Here we have extended this finding. We show by cell/nuclear fractionation followed by microarray analysis that a number of miRNAs can be detected within the nucleolus of rat myoblasts, some of which are significantly concentrated there. Pronounced nucleolar localization is a specific phenomenon since other miRNAs are present at only very low levels in the nucleolus and occur at much higher levels in the nucleoplasm and/or the cytoplasm. We have further characterized a subset of these miRNAs using RT-qPCR and in situ hybridization, and the results suggest that some miRNAs are present in the nucleolus in precursor form while others are present as mature species. Furthermore, we have found that these miRNAs are clustered in specific sites within the nucleolus that correspond to the classical granular component. One of these miRNAs is completely homologous to a portion of a snoRNA, suggesting that it may be processed from it. In contrast, the other nucleolar-concentrated miRNAs do not show homology with any annotated rat snoRNAs and thus appear to be present in the nucleolus for other reasons, such as modification/processing, or to play roles in the late stages of ribosome biosynthesis or in nonribosomal functions that have recently been ascribed to the granular component of the nucleolus.
Figures

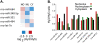

Similar articles
-
MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells.Proc Natl Acad Sci U S A. 2006 Dec 12;103(50):18957-62. doi: 10.1073/pnas.0609466103. Epub 2006 Nov 29. Proc Natl Acad Sci U S A. 2006. PMID: 17135348 Free PMC article.
-
A mRNA and cognate microRNAs localize in the nucleolus.Nucleus. 2014;5(6):636-42. doi: 10.4161/19491034.2014.990864. Nucleus. 2014. PMID: 25485975 Free PMC article.
-
Dynamic localisation of mature microRNAs in Human nucleoli is influenced by exogenous genetic materials.PLoS One. 2013 Aug 6;8(8):e70869. doi: 10.1371/journal.pone.0070869. Print 2013. PLoS One. 2013. PMID: 23940654 Free PMC article.
-
Beyond ribosome biogenesis: noncoding nucleolar RNAs in physiology and tumor biology.Nucleus. 2023 Dec;14(1):2274655. doi: 10.1080/19491034.2023.2274655. Epub 2023 Oct 31. Nucleus. 2023. PMID: 37906621 Free PMC article. Review.
-
Localization of ribosomal protein S1 in the granular component of the interphase nucleolus and its distribution during mitosis.J Cell Biol. 1985 Mar;100(3):873-86. doi: 10.1083/jcb.100.3.873. J Cell Biol. 1985. PMID: 3882724 Free PMC article. Review.
Cited by
-
New insights into nucleolar structure and function.F1000Prime Rep. 2015 Apr 2;7:48. doi: 10.12703/P7-48. eCollection 2015. F1000Prime Rep. 2015. PMID: 26097721 Free PMC article. Review.
-
Nuclear miR-30b-5p suppresses TFEB-mediated lysosomal biogenesis and autophagy.Cell Death Differ. 2021 Jan;28(1):320-336. doi: 10.1038/s41418-020-0602-4. Epub 2020 Aug 6. Cell Death Differ. 2021. PMID: 32764647 Free PMC article.
-
The Cajal body and the nucleolus: "In a relationship" or "It's complicated"?RNA Biol. 2017 Jun 3;14(6):739-751. doi: 10.1080/15476286.2016.1236169. Epub 2016 Sep 23. RNA Biol. 2017. PMID: 27661468 Free PMC article. Review.
-
Noncoding RNAs link metabolic reprogramming to immune microenvironment in cancers.J Hematol Oncol. 2021 Oct 15;14(1):169. doi: 10.1186/s13045-021-01179-y. J Hematol Oncol. 2021. PMID: 34654454 Free PMC article. Review.
-
A comprehensive characterization of the nuclear microRNA repertoire of post-mitotic neurons.Front Mol Neurosci. 2013 Nov 26;6:43. doi: 10.3389/fnmol.2013.00043. eCollection 2013. Front Mol Neurosci. 2013. PMID: 24324399 Free PMC article.
References
-
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002;12:1–11. - PubMed
-
- Bártová E, Harnicarová A, Krejcí J, Strasák L, Kozubek S. Single-cell c-myc gene expression in relationship to nuclear domains. Chromosome Res. 2008;16:325–343. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
