Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing
- PMID: 19628622
- PMCID: PMC2743053
- DOI: 10.1261/rna.1724409
Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing
Abstract
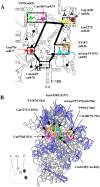
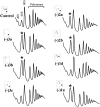
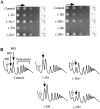
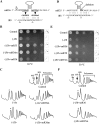
The ribosome decoding center is rich in modified rRNA nucleotides and little is known about their effects. Here, we examine the consequences of systematically deleting eight pseudouridine and 2'-O-methylation modifications in the yeast decoding center. Loss of most modifications individually has no apparent effect on cell growth. However, deletions of 2-3 modifications in the A- and P-site regions can cause (1) reduced growth rates (approximately 15%-50% slower); (2) reduced amino acid incorporation rates (14%-24% slower); and (3) a significant deficiency in free small subunits. Negative and positive interference effects were observed, as well as strong positional influences. Notably, blocking formation of a hypermodified pseudouridine in the P region delays the onset of the final cleavage event in 18S rRNA formation ( approximately 60% slower), suggesting that modification at this site could have an important role in modulating ribosome synthesis.
Figures



References
-
- Bakin A, Ofengand J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyl transferase center: Analysis by the application of a new sequencing technique. Biochemistry. 1993;32:9754–9762. - PubMed
-
- Brand RC, Klootwijk J, Van Steenbergen TJ, De Kok AJ, Planta RJ. Secondary methylation of yeast ribosomal precursor RNA. Eur J Biochem. 1977;75:311–318. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
