Triplex structures in an RNA pseudoknot enhance mechanical stability and increase efficiency of -1 ribosomal frameshifting
- PMID: 19628688
- PMCID: PMC2722267
- DOI: 10.1073/pnas.0905046106
Triplex structures in an RNA pseudoknot enhance mechanical stability and increase efficiency of -1 ribosomal frameshifting
Abstract
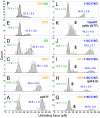
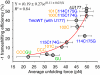
Many viruses use programmed -1 ribosomal frameshifting to express defined ratios of structural and enzymatic proteins. Pseudoknot structures in messenger RNAs stimulate frameshifting in upstream slippery sequences. The detailed molecular determinants of pseudoknot mechanical stability and frameshifting efficiency are not well understood. Here we use single-molecule unfolding studies by optical tweezers, and frameshifting assays to elucidate how mechanical stability of a pseudoknot and its frameshifting efficiency are regulated by tertiary stem-loop interactions. Mechanical unfolding of a model pseudoknot and mutants designed to dissect specific interactions reveals that mechanical stability depends strongly on triplex structures formed by stem-loop interactions. Combining single-molecule and mutational studies facilitates the identification of pseudoknot folding intermediates. Average unfolding forces of the pseudoknot and mutants ranging from 50 to 22 picoNewtons correlated with frameshifting efficiencies ranging from 53% to 0%. Formation of major-groove and minor-groove triplex structures enhances pseudoknot stem stability and torsional resistance, and may thereby stimulate frameshifting. Better understanding of the molecular determinants of frameshifting efficiency may facilitate the development of anti-virus therapeutics targeting frameshifting.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

