The BRC repeats of human BRCA2 differentially regulate RAD51 binding on single- versus double-stranded DNA to stimulate strand exchange
- PMID: 19628690
- PMCID: PMC2714763
- DOI: 10.1073/pnas.0906208106
The BRC repeats of human BRCA2 differentially regulate RAD51 binding on single- versus double-stranded DNA to stimulate strand exchange
Abstract
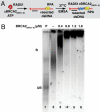
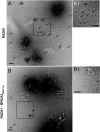
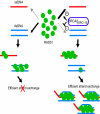
The breast and ovarian cancer suppressor BRCA2 controls the enzyme RAD51 during homologous DNA recombination (HDR) to preserve genome stability. BRCA2 binds to RAD51 through 8 conserved BRC repeat motifs dispersed in an 1127-residue region (BRCA2([BRC1-8])). Here, we show that BRCA2([BRC1-8]) exerts opposing effects on the binding of RAD51 to single-stranded (ss) versus double-stranded (ds) DNA substrates, enhancing strand exchange. BRCA2([BRC1-8]) alters the electrophoretic mobility of RAD51 bound to an ssDNA substrate, accompanied by an increase in ssDNA-bound protein assemblies, revealed by electron microscopy. Single-molecule fluorescence spectroscopy shows that BRCA2([BRC1-8]) promotes RAD51 loading onto ssDNA. In contrast, BRCA2([BRC1-8]) has a different effect on RAD51 assembly on dsDNA; it suppresses and slows this process. When homologous ssDNA and dsDNA are both present, BRCA2([BRC1-8]) stimulates strand exchange, with delayed RAD51 loading onto dsDNA accompanying the appearance of joint molecules representing recombination products. Collectively, our findings suggest that BRCA2([BRC1-8]) targets RAD51 to ssDNA while inhibiting dsDNA binding and that these contrasting activities together bolster one another to stimulate HDR. Our work provides fresh insight into the mechanism of HDR in humans, and its regulation by the BRCA2 tumor suppressor.
Conflict of interest statement
Conflict of interest statement: The editor and A.V. recently (March 2009) co-authored a paper on a related topic.
Figures



References
-
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. - PubMed
-
- Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–272. - PubMed
-
- Thorslund T, West SC. BRCA2: A universal recombinase regulator. Oncogene. 2007;26:7720–7730. - PubMed
-
- Wong AK, Pero R, Ormonde PA, Tavtigian SV, Bartel PL. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J Biol Chem. 1997;272:31941–31944. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

