Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates
- PMID: 19628699
- PMCID: PMC2714764
- DOI: 10.1073/pnas.0906413106
Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates
Abstract
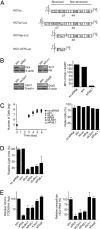
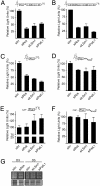
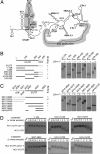
Inevitably, viruses depend on host factors for their multiplication. Here, we show that hepatitis C virus (HCV) RNA translation and replication depends on Rck/p54, LSm1, and PatL1, which regulate the fate of cellular mRNAs from translation to degradation in the 5'-3'-deadenylation-dependent mRNA decay pathway. The requirement of these proteins for efficient HCV RNA translation was linked to the 5' and 3' untranslated regions (UTRs) of the viral genome. Furthermore, LSm1-7 complexes specifically interacted with essential cis-acting HCV RNA elements located in the UTRs. These results bridge HCV life cycle requirements and highly conserved host proteins of cellular mRNA decay. The previously described role of these proteins in the replication of 2 other positive-strand RNA viruses, the plant brome mosaic virus and the bacteriophage Qss, pinpoint a weak spot that may be exploited to generate broad-spectrum antiviral drugs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

