DNA polymerase epsilon, acetylases and remodellers cooperate to form a specialized chromatin structure at a tRNA insulator
- PMID: 19629037
- PMCID: PMC2738695
- DOI: 10.1038/emboj.2009.198
DNA polymerase epsilon, acetylases and remodellers cooperate to form a specialized chromatin structure at a tRNA insulator
Abstract
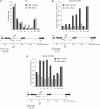
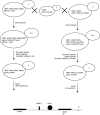
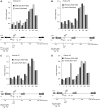
Insulators bind transcription factors and use chromatin remodellers and modifiers to mediate insulation. In this report, we identified proteins required for the efficient formation and maintenance of a specialized chromatin structure at the yeast tRNA insulator. The histone acetylases, SAS-I and NuA4, functioned in insulation, independently of tRNA and did not participate in the formation of the hypersensitive site at the tRNA. In contrast, DNA polymerase epsilon, functioned with the chromatin remodeller, Rsc, and the histone acetylase, Rtt109, to generate a histone-depleted region at the tRNA insulator. Rsc and Rtt109 were required for efficient binding of TFIIIB to the tRNA insulator, and the bound transcription factor and Rtt109 in turn were required for the binding of Rsc to tRNA. Robust insulation during growth and cell division involves the formation of a hypersensitive site at the insulator during chromatin maturation together with competition between acetylases and deacetylases.
Figures
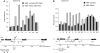




Similar articles
-
Transcription independent insulation at TFIIIC-dependent insulators.Genetics. 2009 Sep;183(1):131-48. doi: 10.1534/genetics.109.106203. Epub 2009 Jul 13. Genetics. 2009. PMID: 19596900 Free PMC article.
-
Silencing near tRNA genes is nucleosome-mediated and distinct from boundary element function.Gene. 2013 Aug 15;526(1):7-15. doi: 10.1016/j.gene.2013.05.016. Epub 2013 May 23. Gene. 2013. PMID: 23707796 Free PMC article.
-
Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14.EMBO J. 2004 Mar 24;23(6):1348-59. doi: 10.1038/sj.emboj.7600143. Epub 2004 Mar 4. EMBO J. 2004. PMID: 15014446 Free PMC article.
-
ISWI complexes in Saccharomyces cerevisiae.Biochim Biophys Acta. 2004 Mar 15;1677(1-3):100-12. doi: 10.1016/j.bbaexp.2003.10.014. Biochim Biophys Acta. 2004. PMID: 15020051 Review.
-
Gene insulation. Part I: natural strategies in yeast and Drosophila.Biochem Cell Biol. 2010 Dec;88(6):875-84. doi: 10.1139/O10-110. Biochem Cell Biol. 2010. PMID: 21102650 Review.
Cited by
-
Global regulation of heterochromatin spreading by Leo1.Open Biol. 2015 May;5(5):150045. doi: 10.1098/rsob.150045. Open Biol. 2015. PMID: 25972440 Free PMC article.
-
Dynamics of Sir3 spreading in budding yeast: secondary recruitment sites and euchromatic localization.EMBO J. 2011 Mar 16;30(6):1012-26. doi: 10.1038/emboj.2011.30. Epub 2011 Feb 18. EMBO J. 2011. PMID: 21336256 Free PMC article.
-
Long-range heterochromatin association is mediated by silencing and double-strand DNA break repair proteins.J Cell Biol. 2013 Jun 10;201(6):809-26. doi: 10.1083/jcb.201211105. Epub 2013 Jun 3. J Cell Biol. 2013. PMID: 23733345 Free PMC article.
-
tRNA Genes Affect Chromosome Structure and Function via Local Effects.Mol Cell Biol. 2019 Apr 2;39(8):e00432-18. doi: 10.1128/MCB.00432-18. Print 2019 Apr 15. Mol Cell Biol. 2019. PMID: 30718362 Free PMC article.
-
COMPASS and SWI/SNF complexes in development and disease.Nat Rev Genet. 2021 Jan;22(1):38-58. doi: 10.1038/s41576-020-0278-0. Epub 2020 Sep 21. Nat Rev Genet. 2021. PMID: 32958894 Review.
References
-
- Adkins MW, Howar SR, Tyler JK (2004) Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol Cell 14: 657–666 - PubMed
-
- Annunziato AT, Schindler RK, Thomas CA Jr, Seale RL (1981) Dual nature of newly replicated chromatin. Evidence for nucleosomal and non-nucleosomal DNA at the site of native replication forks. J Biol Chem 256: 11880–11886 - PubMed
-
- Annunziato AT, Seale RL (1983) Histone deacetylation is required for the maturation of newly replicated chromatin. J Biol Chem 258: 12675–12684 - PubMed
-
- Asturias FJ, Cheung IK, Sabouri N, Chilkova O, Wepplo D, Johansson E (2006) Structure of Saccharomyces cerevisiae DNA polymerase epsilon by cryo-electron microscopy. Nat Struct Mol Biol 13: 35–43 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

