Silent chromatin at the middle and ends: lessons from yeasts
- PMID: 19629038
- PMCID: PMC2722250
- DOI: 10.1038/emboj.2009.185
Silent chromatin at the middle and ends: lessons from yeasts
Abstract
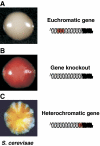
Eukaryotic centromeres and telomeres are specialized chromosomal regions that share one common characteristic: their underlying DNA sequences are assembled into heritably repressed chromatin. Silent chromatin in budding and fission yeast is composed of fundamentally divergent proteins tat assemble very different chromatin structures. However, the ultimate behaviour of silent chromatin and the pathways that assemble it seem strikingly similar among Saccharomyces cerevisiae (S. cerevisiae), Schizosaccharomyces pombe (S. pombe) and other eukaryotes. Thus, studies in both yeasts have been instrumental in dissecting the mechanisms that establish and maintain silent chromatin in eukaryotes, contributing substantially to our understanding of epigenetic processes. In this review, we discuss current models for the generation of heterochromatic domains at centromeres and telomeres in the two yeast species.
Figures
References
-
- Akhtar A, Gasser SM (2007) The nuclear envelope and transcriptional control. Nat Rev Genet 8: 507–517 - PubMed
-
- Alfredsson-Timmins J, Henningson F, Bjerling P (2007) The Clr4 methyltransferase determines the subnuclear localization of the mating-type region in fission yeast. J Cell Sci 120 (Pt 11): 1935–1943 - PubMed
-
- Allshire RC, Javerzat JP, Redhead NJ, Cranston G (1994) Position effect variegation at fission yeast centromeres. Cell 76: 157–169 - PubMed
-
- Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G (1995) Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev 9: 218–233 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

