Taming the tiger by the tail: modulation of DNA damage responses by telomeres
- PMID: 19629039
- PMCID: PMC2722249
- DOI: 10.1038/emboj.2009.176
Taming the tiger by the tail: modulation of DNA damage responses by telomeres
Abstract
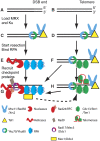
Telomeres are by definition stable and inert chromosome ends, whereas internal chromosome breaks are potent stimulators of the DNA damage response (DDR). Telomeres do not, as might be expected, exclude DDR proteins from chromosome ends but instead engage with many DDR proteins. However, the most powerful DDRs, those that might induce chromosome fusion or cell-cycle arrest, are inhibited at telomeres. In budding yeast, many DDR proteins that accumulate most rapidly at double strand breaks (DSBs), have important functions in physiological telomere maintenance, whereas DDR proteins that arrive later tend to have less important functions. Considerable diversity in telomere structure has evolved in different organisms and, perhaps reflecting this diversity, different DDR proteins seem to have distinct roles in telomere physiology in different organisms. Drawing principally on studies in simple model organisms such as budding yeast, in which many fundamental aspects of the DDR and telomere biology have been established; current views on how telomeres harness aspects of DDR pathways to maintain telomere stability and permit cell-cycle division are discussed.
Figures

Similar articles
-
Similarities and differences between "uncapped" telomeres and DNA double-strand breaks.Chromosoma. 2012 Apr;121(2):117-30. doi: 10.1007/s00412-011-0357-2. Epub 2011 Dec 28. Chromosoma. 2012. PMID: 22203190 Review.
-
Hiding at the ends of yeast chromosomes: telomeres, nucleases and checkpoint pathways.J Cell Sci. 2003 Oct 15;116(Pt 20):4057-65. doi: 10.1242/jcs.00765. J Cell Sci. 2003. PMID: 12972499 Review.
-
Drosophila cell cycle under arrest: uncapped telomeres plead guilty.Cell Cycle. 2009 Apr 1;8(7):990-5. doi: 10.4161/cc.8.7.7960. Epub 2009 Apr 4. Cell Cycle. 2009. PMID: 19270525
-
Functional links between telomeres and proteins of the DNA-damage response.Genes Dev. 2004 Aug 1;18(15):1781-99. doi: 10.1101/gad.1214504. Genes Dev. 2004. PMID: 15289453 Review.
-
The regulation of the DNA damage response at telomeres: focus on kinases.Biochem Soc Trans. 2021 Apr 30;49(2):933-943. doi: 10.1042/BST20200856. Biochem Soc Trans. 2021. PMID: 33769480 Review.
Cited by
-
The Telomere Binding Protein Cdc13 and the Single-Stranded DNA Binding Protein RPA Protect Telomeric DNA from Resection by Exonucleases.J Mol Biol. 2015 Sep 25;427(19):3023-30. doi: 10.1016/j.jmb.2015.08.002. Epub 2015 Aug 8. J Mol Biol. 2015. PMID: 26264873 Free PMC article.
-
A CDK-Dependent Phosphorylation of a Novel Domain of Rif1 Regulates its Function during Telomere Damage and Other Types of Stress.Mol Cell Biol. 2023;43(5):185-199. doi: 10.1080/10985549.2023.2193768. Epub 2023 May 4. Mol Cell Biol. 2023. PMID: 37140180 Free PMC article.
-
Vps74 Connects the Golgi Apparatus and Telomeres in Saccharomyces cerevisiae.G3 (Bethesda). 2018 May 4;8(5):1807-1816. doi: 10.1534/g3.118.200172. G3 (Bethesda). 2018. PMID: 29593073 Free PMC article.
-
Condensin II subunit NCAPH2 associates with shelterin protein TRF1 and is required for telomere stability.J Cell Physiol. 2019 Nov;234(11):20755-20768. doi: 10.1002/jcp.28681. Epub 2019 Apr 26. J Cell Physiol. 2019. PMID: 31026066 Free PMC article.
-
DNA-end capping by the budding yeast transcription factor and subtelomeric binding protein Tbf1.EMBO J. 2012 Jan 4;31(1):138-49. doi: 10.1038/emboj.2011.349. Epub 2011 Sep 27. EMBO J. 2012. PMID: 21952045 Free PMC article.
References
-
- Addinall SG, Downey M, Yu M, Zubko MK, Dewar J, Leake A, Hallinan J, Shaw O, James K, Wilkinson DJ, Wipat A, Durocher D, Lydall D (2008) A genomewide suppressor and enhancer analysis of cdc13-1 reveals varied cellular processes influencing telomere capping in Saccharomyces cerevisiae. Genetics 180: 2251–2266 - PMC - PubMed
-
- Ahmed S, Hodgkin J (2000) MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans [see comments]. Nature 403: 159–164 - PubMed
-
- Artandi SE, Attardi LD (2005) Pathways connecting telomeres and p53 in senescence, apoptosis, and cancer. Biochem Biophys Res Commun 331: 881–890 - PubMed
-
- Aubert G, Lansdorp PM (2008) Telomeres and aging. Physiol Rev 88: 557–579 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

