Focus on the centre: the role of chromatin on the regulation of centromere identity and function
- PMID: 19629040
- PMCID: PMC2722248
- DOI: 10.1038/emboj.2009.174
Focus on the centre: the role of chromatin on the regulation of centromere identity and function
Abstract
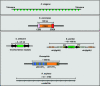
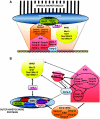
The centromere is a specialised chromosomal structure that regulates faithful chromosome segregation during cell division, as it dictates the site of assembly of the kinetochore, a critical structure that mediates binding of chromosomes to the spindle, monitors bipolar attachment and pulls chromosomes to the poles during anaphase. Identified more than a century ago as the primary constriction of condensed metaphase chromosomes, the centromere remained elusive to molecular characterisation for many years owed to its unusual enrichment in highly repetitive satellite DNA sequences, except in budding yeast. In the last decade, our understanding of centromere structure, organisation and function has increased tremendously. Nowadays, we know that centromere identity is determined epigenetically by the formation of a unique type of chromatin, which is characterised by the presence of the centromere-specific histone H3 variant CenH3, originally called CENP-A, which replaces canonical histone H3 at centromeres. CenH3-chromatin constitutes the physical and functional foundation for kinetochore assembly. This review explores recent studies addressing the structural and functional characterisation of CenH3-chromatin, its assembly and propagation during mitosis, and its contribution to kinetochore assembly.
Figures



Similar articles
-
In vitro centromere and kinetochore assembly on defined chromatin templates.Nature. 2011 Aug 28;477(7364):354-8. doi: 10.1038/nature10379. Nature. 2011. PMID: 21874020 Free PMC article.
-
Epigenomics of centromere assembly and function.Curr Opin Cell Biol. 2010 Dec;22(6):772-80. doi: 10.1016/j.ceb.2010.07.002. Epub 2010 Jul 31. Curr Opin Cell Biol. 2010. PMID: 20675111 Review.
-
Nucleosomal composition at the centromere: a numbers game.Chromosome Res. 2013 Mar;21(1):27-36. doi: 10.1007/s10577-012-9335-7. Epub 2013 Jan 18. Chromosome Res. 2013. PMID: 23328870 Free PMC article. Review.
-
Ectopic centromere nucleation by CENP--a in fission yeast.Genetics. 2014 Dec;198(4):1433-46. doi: 10.1534/genetics.114.171173. Epub 2014 Oct 7. Genetics. 2014. PMID: 25298518 Free PMC article.
-
Structural determinants for generating centromeric chromatin.Nature. 2004 Jul 29;430(6999):578-82. doi: 10.1038/nature02766. Nature. 2004. PMID: 15282608
Cited by
-
Drosophila CENP-C is essential for centromere identity.Chromosoma. 2011 Feb;120(1):83-96. doi: 10.1007/s00412-010-0293-6. Epub 2010 Sep 23. Chromosoma. 2011. PMID: 20862486
-
Deposition, turnover, and release of CENH3 at Arabidopsis centromeres.Chromosoma. 2011 Dec;120(6):633-40. doi: 10.1007/s00412-011-0338-5. Epub 2011 Aug 13. Chromosoma. 2011. PMID: 21842230
-
The case for junk DNA.PLoS Genet. 2014 May 8;10(5):e1004351. doi: 10.1371/journal.pgen.1004351. eCollection 2014 May. PLoS Genet. 2014. PMID: 24809441 Free PMC article. No abstract available.
-
Diatom centromeres suggest a mechanism for nuclear DNA acquisition.Proc Natl Acad Sci U S A. 2017 Jul 18;114(29):E6015-E6024. doi: 10.1073/pnas.1700764114. Epub 2017 Jul 3. Proc Natl Acad Sci U S A. 2017. PMID: 28673987 Free PMC article.
-
Genetic insights into non-obstructive azoospermia: Implications for diagnosis and TESE outcomes.J Assist Reprod Genet. 2025 Apr;42(4):1223-1237. doi: 10.1007/s10815-025-03409-5. Epub 2025 Feb 11. J Assist Reprod Genet. 2025. PMID: 39932629 Free PMC article.
References
-
- Agudo M, Abad JP, Molina I, Losada A, Ripoll P, Villasante A (2000) A dicentric chromosome of Drosophila melanogaster showing alternate centromere inactivation. Chromosoma 109: 190–196 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

