Harvesting candidate genes responsible for serious adverse drug reactions from a chemical-protein interactome
- PMID: 19629158
- PMCID: PMC2704868
- DOI: 10.1371/journal.pcbi.1000441
Harvesting candidate genes responsible for serious adverse drug reactions from a chemical-protein interactome
Abstract
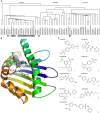
Identifying genetic factors responsible for serious adverse drug reaction (SADR) is of critical importance to personalized medicine. However, genome-wide association studies are hampered due to the lack of case-control samples, and the selection of candidate genes is limited by the lack of understanding of the underlying mechanisms of SADRs. We hypothesize that drugs causing the same type of SADR might share a common mechanism by targeting unexpectedly the same SADR-mediating protein. Hence we propose an approach of identifying the common SADR-targets through constructing and mining an in silico chemical-protein interactome (CPI), a matrix of binding strengths among 162 drug molecules known to cause at least one type of SADR and 845 proteins. Drugs sharing the same SADR outcome were also found to possess similarities in their CPI profiles towards this 845 protein set. This methodology identified the candidate gene of sulfonamide-induced toxic epidermal necrolysis (TEN): all nine sulfonamides that cause TEN were found to bind strongly to MHC I (Cw*4), whereas none of the 17 control drugs that do not cause TEN were found to bind to it. Through an insight into the CPI, we found the Y116S substitution of MHC I (B*5703) enhances the unexpected binding of abacavir to its antigen presentation groove, which explains why B*5701, not B*5703, is the risk allele of abacavir-induced hypersensitivity. In conclusion, SADR targets and the patient-specific off-targets could be identified through a systematic investigation of the CPI, generating important hypotheses for prospective experimental validation of the candidate genes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
SePreSA: a server for the prediction of populations susceptible to serious adverse drug reactions implementing the methodology of a chemical-protein interactome.Nucleic Acids Res. 2009 Jul;37(Web Server issue):W406-12. doi: 10.1093/nar/gkp312. Epub 2009 May 5. Nucleic Acids Res. 2009. PMID: 19417066 Free PMC article.
-
Identifying unexpected therapeutic targets via chemical-protein interactome.PLoS One. 2010 Mar 8;5(3):e9568. doi: 10.1371/journal.pone.0009568. PLoS One. 2010. PMID: 20221449 Free PMC article.
-
Chemical-protein interactome and its application in off-target identification.Interdiscip Sci. 2011 Mar;3(1):22-30. doi: 10.1007/s12539-011-0051-8. Epub 2011 Mar 3. Interdiscip Sci. 2011. PMID: 21369884 Review.
-
DPDR-CPI, a server that predicts Drug Positioning and Drug Repositioning via Chemical-Protein Interactome.Sci Rep. 2016 Nov 2;6:35996. doi: 10.1038/srep35996. Sci Rep. 2016. PMID: 27805045 Free PMC article.
-
Data-driven methods to discover molecular determinants of serious adverse drug events.Clin Pharmacol Ther. 2009 Mar;85(3):259-68. doi: 10.1038/clpt.2008.274. Epub 2009 Jan 28. Clin Pharmacol Ther. 2009. PMID: 19177064 Free PMC article. Review.
Cited by
-
Binding of protein kinase inhibitors to synapsin I inferred from pair-wise binding site similarity measurements.PLoS One. 2010 Aug 16;5(8):e12214. doi: 10.1371/journal.pone.0012214. PLoS One. 2010. PMID: 20808948 Free PMC article.
-
Molecular Docking: Shifting Paradigms in Drug Discovery.Int J Mol Sci. 2019 Sep 4;20(18):4331. doi: 10.3390/ijms20184331. Int J Mol Sci. 2019. PMID: 31487867 Free PMC article. Review.
-
Structure and dynamics of molecular networks: a novel paradigm of drug discovery: a comprehensive review.Pharmacol Ther. 2013 Jun;138(3):333-408. doi: 10.1016/j.pharmthera.2013.01.016. Epub 2013 Feb 4. Pharmacol Ther. 2013. PMID: 23384594 Free PMC article. Review.
-
Molecular Docking Analysis of Four Drugs (Phenytoin, Amoxicillin, Aceclofenac and Ciprofloxacin) and Their Association With Four Human Leukocyte Antigen (HLA) Alleles.Cureus. 2024 Jun 12;16(6):e62269. doi: 10.7759/cureus.62269. eCollection 2024 Jun. Cureus. 2024. PMID: 39006565 Free PMC article.
-
Molecular docking as a popular tool in drug design, an in silico travel.Adv Appl Bioinform Chem. 2016 Jun 28;9:1-11. doi: 10.2147/AABC.S105289. eCollection 2016. Adv Appl Bioinform Chem. 2016. PMID: 27390530 Free PMC article. Review.
References
-
- Need AC, Motulsky AG, Goldstein DB. Priorities and standards in pharmacogenetic research. Nat Genet. 2005;37:671–681. - PubMed
-
- Tan S, Hall IP, Dewar J, Dow E, Lipworth B. Association between beta 2-adrenoceptor polymorphism and susceptibility to bronchodilator desensitisation in moderately severe stable asthmatics. Lancet. 1997;350:995–999. - PubMed
-
- Fiegenbaum M, da Silveira FR, Van der Sand CR, Van der Sand LC, Ferreira ME, et al. The role of common variants of ABCB1, CYP3A4, and CYP3A5 genes in lipid-lowering efficacy and safety of simvastatin treatment. Clin Pharmacol Ther. 2005;78:551–558. - PubMed
-
- Pirmohamed M, Park BK. Genetic susceptibility to adverse drug reactions. Trends Pharmacol Sci. 2001;22:298–305. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

